Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression
Chenglong Xia*, Jean Fan*, George Emanuel*, Junjie Hao, and Xiaowei Zhuang^
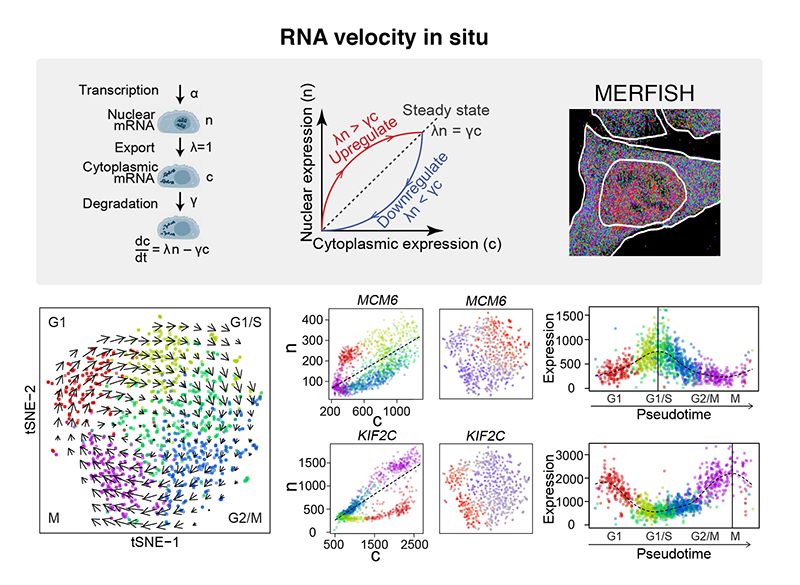
Abstract: The spatial organization of RNAs within cells and spatial patterning of cells within tissues play crucial roles in many biological processes. Here, we demonstrate that multiplexed error-robust FISH (MERFISH) can achieve near-genome-wide, spatially resolved RNA profiling of individual cells with high accuracy and high detection efficiency. Using this approach, we identified RNA species enriched in different subcellular compartments, observed transcriptionally distinct cell states corresponding to different cell-cycle phases, and revealed spatial patterning of transcriptionally distinct cells. Spatially resolved transcriptome quantification within cells further enabled RNA velocity and pseudotime analysis, which revealed numerous genes with cell cycle-dependent expression. We anticipate that spatially resolved transcriptome analysis will advance our understanding of the interplay between gene regulation and spatial context in biological systems.
Paper: PNAS. Sept 9, 2019, doi:10.1073/pnas.1912459116
| Pubmed
| PDF