VeloViz - RNA-velocity informed embeddings for visualizing cellular trajectories
Lyla Atta, Arpan Sahoo, Jean Fan^
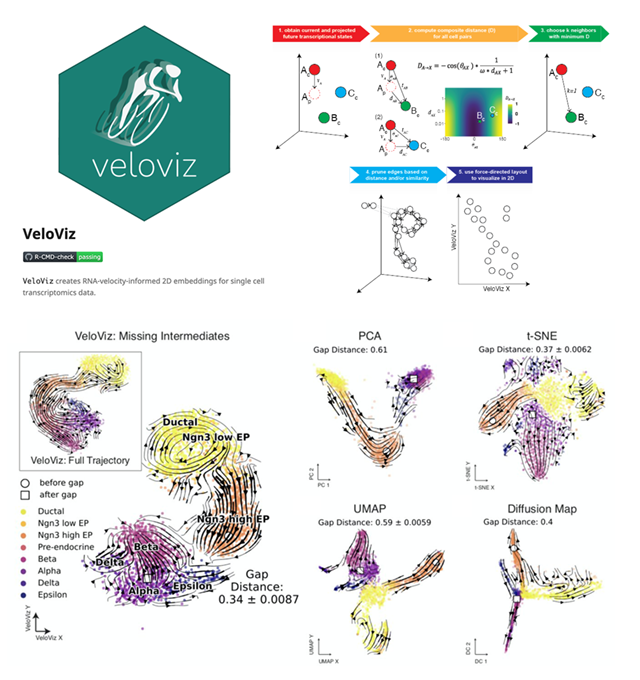
Abstract: Single cell transcriptomic technologies enable genome-wide gene expression measurements in individual cells but can only provide a static snapshot of cell states. RNA velocity analysis can infer cell state changes from single cell transcriptomics data. To interpret these cell state changes as part of underlying cellular trajectories, current approaches rely on visualization with principal components, t-distributed stochastic neighbor embedding, and other 2D embeddings derived from the observed single cell transcriptional states. However, these 2D embeddings can yield different representations of the underlying cellular trajectories, hindering the interpretation of cell state changes. We developed VeloViz to create RNA-velocity-informed 2D and 3D embeddings from single cell transcriptomics data. Using both real and simulated data, we demonstrate that VeloViz embeddings are able to consistently capture underlying cellular trajectories across diverse trajectory topologies, even when intermediate cell states may be missing. By taking into consideration the predicted future transcriptional states from RNA velocity analysis, VeloViz can help visualize a more reliable representation of underlying cellular trajectories.
Paper: Bioinformatics. September 28, 2021. doi.org/10.1093/bioinformatics/btab653
| Pubmed
| PDF
Relevant code: