
HoneyBADGER
HMM-integrated Bayesian approach for detecting CNV and LOH events from single-cell RNA-seq data
HoneyBADGER
HoneyBADGER (hidden Markov model integrated Bayesian approach for detecting CNV and LOH events from single-cell RNA-seq data) identifies and infers the presence of CNV and LOH events in single cells and reconstructs subclonal architecture using allele and expression information from single-cell RNA-sequencing data.
The overall approach is detailed in the following publication:
Fan J*, Lee HO*, Lee S, et al. Linking transcriptional and genetic tumor heterogeneity through allele analysis of single-cell RNA-seq data. Genome Res. 2018;
Benefits and Capabilities
(1) Iterative HMM approach detects CNVs
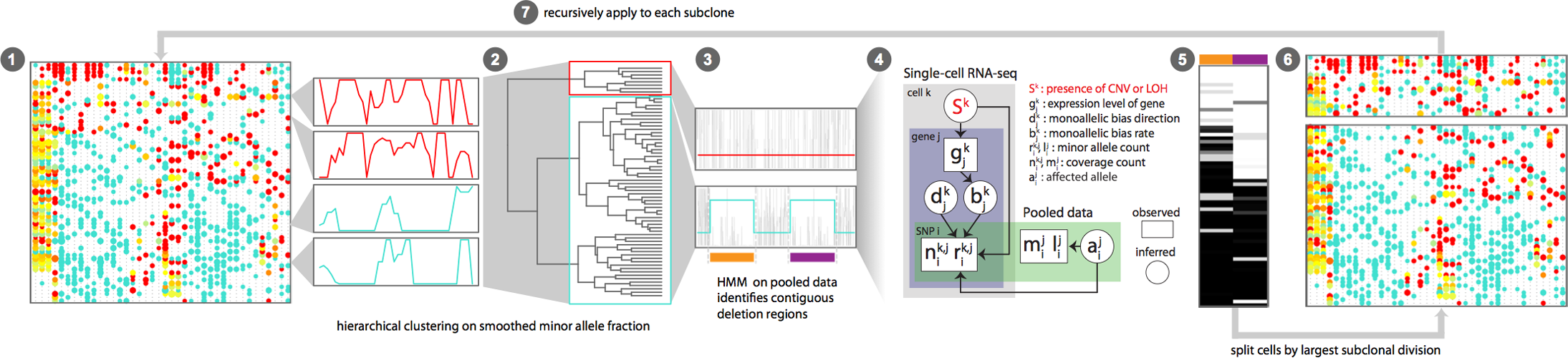
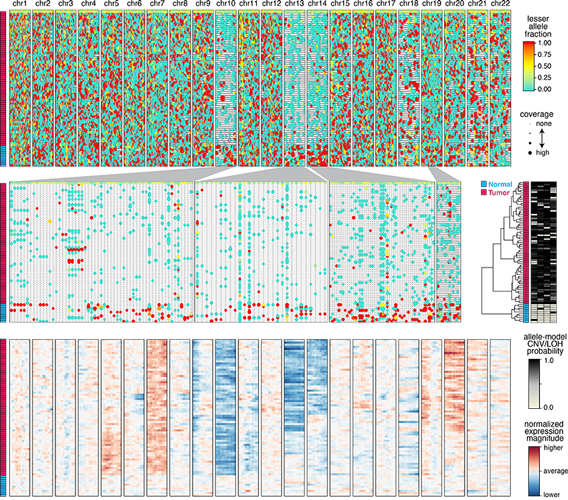
(2) Bayesian hierarchical model uses allele and expression data to infer probability of CNVs in single cells
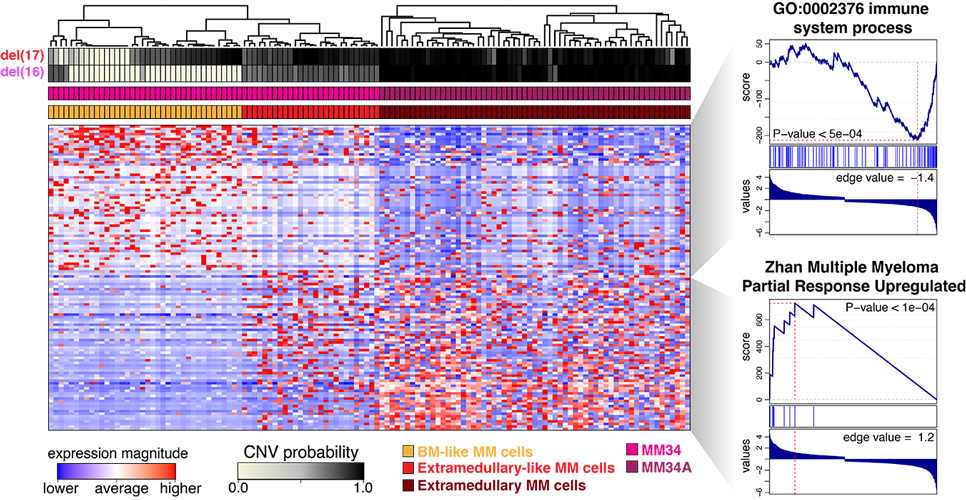
(3) CNV inference from transcriptional data enables transcriptional characterization of subclones and other integrative analyses
Installation
To install HoneyBADGER, we recommend using devtools:
require(devtools)
devtools::install_github('JEFworks/HoneyBADGER')
HoneyBADGER uses JAGS (Just Another Gibbs Sampler) through rjags. Therefore, JAGS must be installed per your operating system requirements. Please see this R-bloggers tutorial for additional tips for installing JAGS and rjags.
Additional dependencies may need to be installed from Bioconductor such as GenomicRanges and others:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("GenomicRanges")
Tutorials
Contributing
We welcome any bug reports, enhancement requests, and other contributions. To submit a bug report or enhancement request, please use the HoneyBADGER GitHub issues tracker. For more substantial contributions, please fork this repo, push your changes to your fork, and submit a pull request with a good commit message.