Analysis of most expressed gene (POSTN): PCA, t-SNE, and UMAP Plots
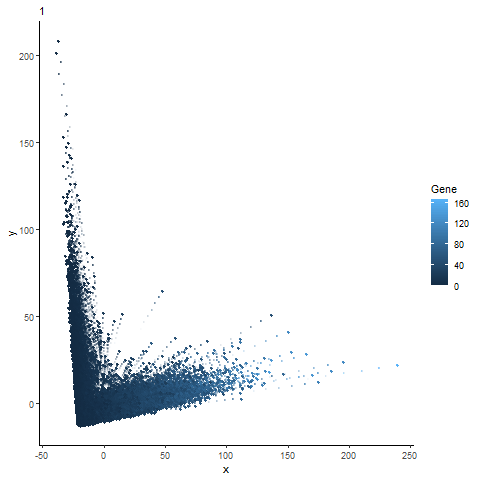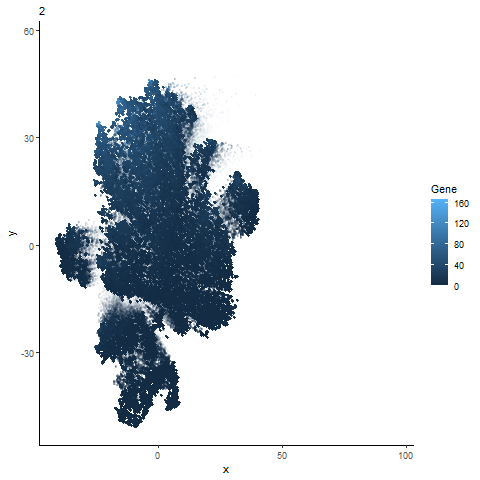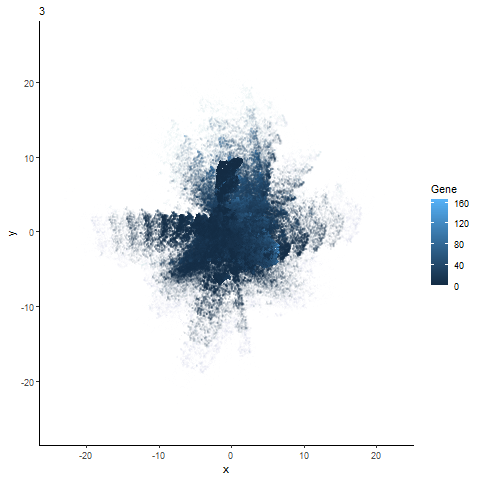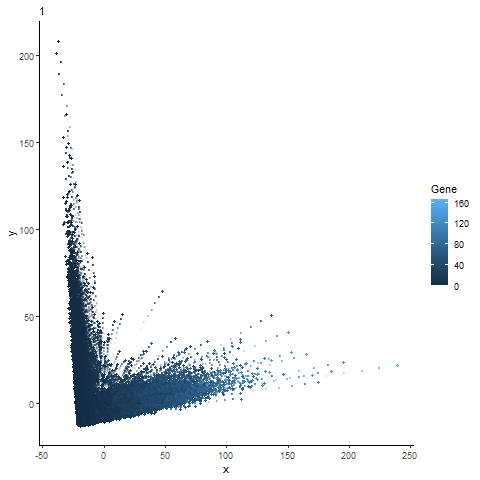
gganimate animation from hw3
Animation of the expression of the top gene POSTN can be expressed across different dimensionality reduction plots (PCA, t-sne, and UMAP).
#### author ####
# Andre Forjaz
# jhed: aperei13
#### load packages ####
library(Rtsne)
library(ggplot2)
library(patchwork)
library(gridExtra)
library(umap)
library(gganimate)
#### Input ####
outpth <-'~/genomic-data-visualization-2024/homework/hwEC1/'
data <- read.csv('~/genomic-data-visualization-2024/data/pikachu.csv.gz', row.names=1)
# data preview
data[1:5,1:8]
# coordinates
coords <- data[,2:3]
# gene expression
gexp <- data[, 6:ncol(data)]
dim(gexp)
colnames(gexp[,1:8])
#### Select top 1 gene ####
topgene <- names(sort(apply(gexp, 2, var), decreasing=TRUE)[1])
topgene
gexpfilter <- gexp[,topgene]
colnames(gexpfilter)
dim(gexpfilter)
#### Run pca ####
pca_res <- prcomp(gexp)
# Make PCA plot
df_top1 <- data.frame(PC1 = pca_res$x[,1],
PC2 = pca_res$x[,2],
Gene = gexpfilter)
colnames(df_top1) <- c('PC1', 'PC2', 'Gene')
p1 <- ggplot(df_top1) +
geom_point(aes(PC1, PC2, color=Gene)) +
scale_color_gradient(low = "black", high = "red",
guide = guide_colorbar(barwidth = 0.7))+
labs(color = topgene, size = 0.5, alpha = 0.6) +
ggtitle("PCA")+
theme_classic()
p1
#### Run tsne ####
tsne_res <- Rtsne(gexp, perplexity = 30, dims = 2)
df_tsne <- data.frame(tSNE1 = tsne_res$Y[, 1],
tSNE2 = tsne_res$Y[, 2],
Gene = gexpfilter)
colnames(df_tsne) <- c('tSNE1', 'tSNE2', 'Gene')
p2 <-ggplot(df_tsne) +
geom_point(aes(tSNE1, tSNE2, color = Gene)) +
scale_color_gradient(low = "black", high = "red",
guide = guide_colorbar(barwidth = 0.7))+
labs(color = topgene, size = 0.5, alpha = 0.6,) +
ggtitle("t-SNE") +
theme_classic()
p2
#### Run UMAP ####
umap_res <- umap(gexp, n_neighbors = 15, n_components = 2)
df_umap <- data.frame(UMAP1 = umap_res$layout[, 1],
UMAP2 = umap_res$layout[, 2],
Gene = gexpfilter)
colnames(df_umap) <- c('UMAP1', 'UMAP2', 'Gene')
p3 <-ggplot(df_umap) +
geom_point(aes(UMAP1 , UMAP2, color = Gene)) +
scale_color_gradient(low = "black", high = "red",
guide = guide_colorbar(barwidth = 0.7))+
labs(color = topgene, size = 0.5, alpha = 0.6,) +
ggtitle("UMAP") +
theme_classic()
p3
#### Plot results ####
p1 + p2 + p3+
plot_annotation(tag_levels = 'a') +
plot_layout(ncol = 3)
combined_plot <-p1 + p2 + p3+
plot_annotation(tag_levels = 'a') +
plot_layout(ncol = 3)
df <- rbind(cbind(df_top1, order=1),
cbind(df_tsne, order=2),
cbind(df_umap, order=3))
head(df)
p <- ggplot(df) +
geom_point(aes(x = x, y = y, col = Gene), size = 1) +
transition_states(order) +
view_follow() +
labs(subtitle="{closest_state}")+
shadow_wake(wake_length = 0.1)+
theme_classic()
animate(p, height = 400, width = 400)
anim_save( paste0(outpth, "hwEC1_aperei13.gif"),p)
#### References ####
# 1- https://rpubs.com/crazyhottommy/pca-in-action
# 2- https://bookdown.org/sjcockell/ismb-tutorial-2023/practical-session-2.html
