Gganimate on HW3 of Varying PC's Influence on tSNEs
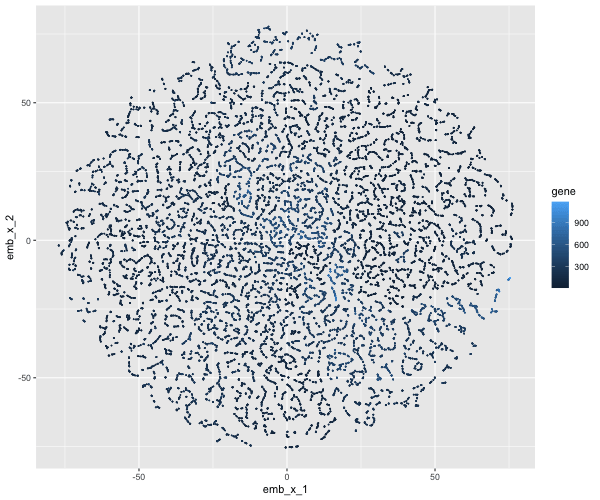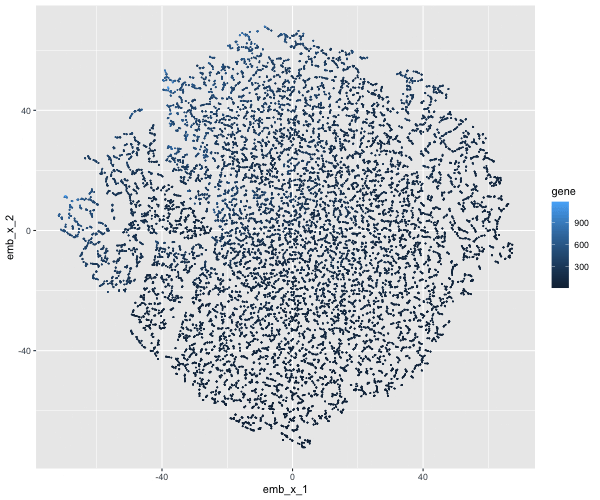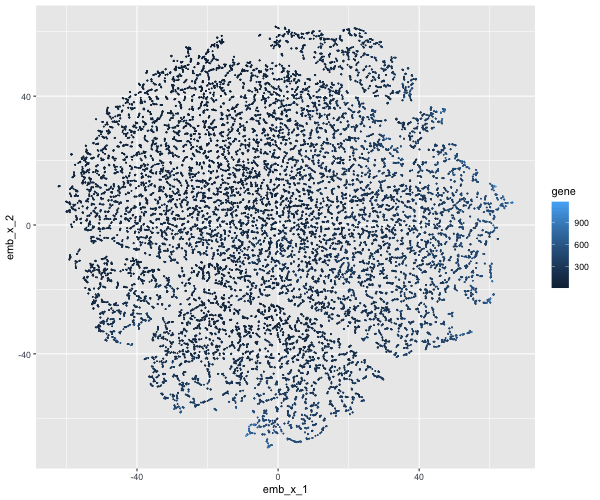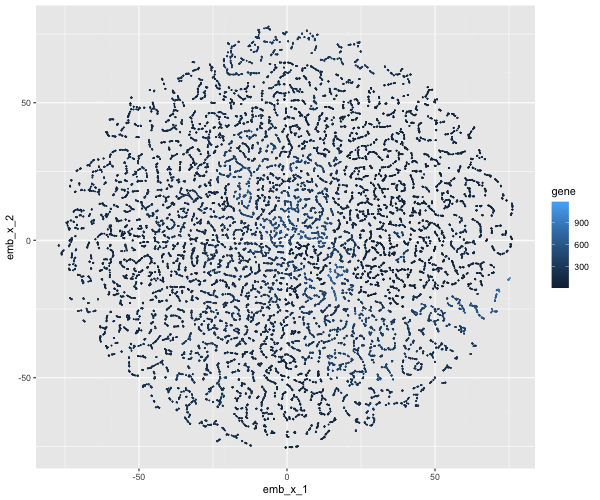
If I perform non-linear dimensionality reduction on PCs, what happens when I vary how many PCs should I use?
If I perform non-linear dimensionality reduction on varying PCs the clustering distinction changes. With less PCs there isn’t defined clusters, but when I add more PCs the clusters become more distinct.
# got saving the code from Caleb
data <- read.csv('~/Documents/genomicsDataVisualization/pikachu.csv.gz', row.names = 1)
library(gganimate)
library(ggplot2)
library(gridExtra)
library(Rtsne)
library(patchwork)
library(here)
gexp <- data[, 6:ncol(data)]
topgene <- names(sort(apply(gexp, 2, var), decreasing=TRUE))
gexpfilter <- gexp[,topgene]
dim(gexpfilter)
pcs <- prcomp(gexpfilter)
# Create a list to store the t-SNE embeddings for different numbers of PCs
emb1 <- Rtsne(pcs$x[,1:2], dims = 2, perplexity = 5)
emb2 <- Rtsne(pcs$x[,1:5], dims = 2, perplexity = 5)
emb3 <- Rtsne(pcs$x[,1:10], dims = 2, perplexity = 5)
emb4 <- Rtsne(pcs$x[,1:15], dims = 2, perplexity = 5)
emb5 <- Rtsne(pcs$x[,1:20], dims = 2, perplexity = 5)
emb6 <- Rtsne(pcs$x[,1:30], dims = 2, perplexity = 5)
#df <- data.frame(pcs$x[,1:2], emb1 = emb1$Y, emb2 = emb2$Y, emb3 = emb3$Y,
# emb4 = emb4$Y, emb5 = emb5$Y, gene = rowSums(gexpfilter))
df1 <- data.frame(pcs$x[,1:2], emb1 = emb1$Y, gene = rowSums(gexpfilter))
colnames(df1) <- c('PC_X', 'PC_Y','emb_x_1', 'emb_x_2', 'gene')
ggplot(df1) + geom_point(aes(x = emb_x_1, emb_x_2, col=gene))
df2 <- data.frame(pcs$x[,1:2], emb2 = emb2$Y, gene = rowSums(gexpfilter))
colnames(df2) <- c('PC_X', 'PC_Y', 'emb_x_1', 'emb_x_2', 'gene')
ggplot(df2) + geom_point(aes(x = emb_x_1, emb_x_2, col=gene))
df3 <- data.frame(pcs$x[,1:2], emb3 = emb3$Y, gene = rowSums(gexpfilter))
colnames(df3) <- c('PC_X', 'PC_Y', 'emb_x_1', 'emb_x_2', 'gene')
ggplot(df3) + geom_point(aes(x = emb_x_1, emb_x_2, col=gene))
df4 <- data.frame(pcs$x[,1:2], emb4 = emb4$Y, gene = rowSums(gexpfilter))
colnames(df4) <- c('PC_X', 'PC_Y', 'emb_x_1', 'emb_x_2', 'gene')
ggplot(df4) + geom_point(aes(x = emb_x_1, emb_x_2, col=gene))
df5 <- data.frame(pcs$x[,1:2], emb5 = emb5$Y, gene = rowSums(gexpfilter))
colnames(df5) <- c('PC_X', 'PC_Y', 'emb_x_1', 'emb_x_2', 'gene')
ggplot(df5) + geom_point(aes(x = emb_x_1, emb_x_2, col=gene))
df6 <- data.frame(pcs$x[,1:2], emb6 = emb6$Y, gene = rowSums(gexpfilter))
colnames(df6) <- c('PC_X', 'PC_Y', 'emb_x_1', 'emb_x_2', 'gene')
ggplot(df6) + geom_point(aes(x = emb_x_1, emb_x_2, col=gene))
df <- rbind(cbind(df1, order=1, size=.1),
cbind(df2, order=2, size=.1),
cbind(df3, order=3, size=0.1),
cbind(df4, order=4, size=.1),
cbind(df5, order=5, size=.1),
cbind(df6, order=6, size=.1))
head(df)
#library(dplyr)
#df <- df %>% select_if(~ !any(is.na(.)))
p <- ggplot(df) + geom_point(aes(x = emb_x_1, y = emb_x_2, col = gene, size = size), size = 0.1)
p
anim <- p + transition_states(order) + view_follow()
# animate
animate(anim, height = 500, width = 600, nframes = 200)
anim_save(here("hwEC1_jmoor185.gif"), animate(anim, height = 500, width = 600, nframes = 200))
