
characterizing spatial gene expression heterogeneity in spatially resolved single-cell transcriptomics data with nonuniform cellular densities
MERINGUE
MERINGUE characterizes spatial gene expression heterogeneity in spatially resolved single-cell transcriptomics data with non-uniform cellular densities.
The overall approach is detailed in the following publication: Miller, B., Bambah-Mukku, D., Dulac, C., Zhuang, X. and Fan, J. Characterizing spatial gene expression heterogeneity in spatially resolved single-cell transcriptomics data with nonuniform cellular densities. Genome Research. May 2021.
Overview
MERINGUE is a computational framework based on spatial auto-correlation and cross-correlation analysis.
You can use MERINGUE to:
- Identify genes with spatially heterogeneous expression
- Group significantly spatially variable genes into primary spatial gene expression patterns
- Identify pairs of genes with complementary expression patterns in spatially co-localized cell-types that may be indicative of cell-cell communication
- Integrate density-agnostic spatial distance weighting to perform spatially-informed transcriptional clustering analysis
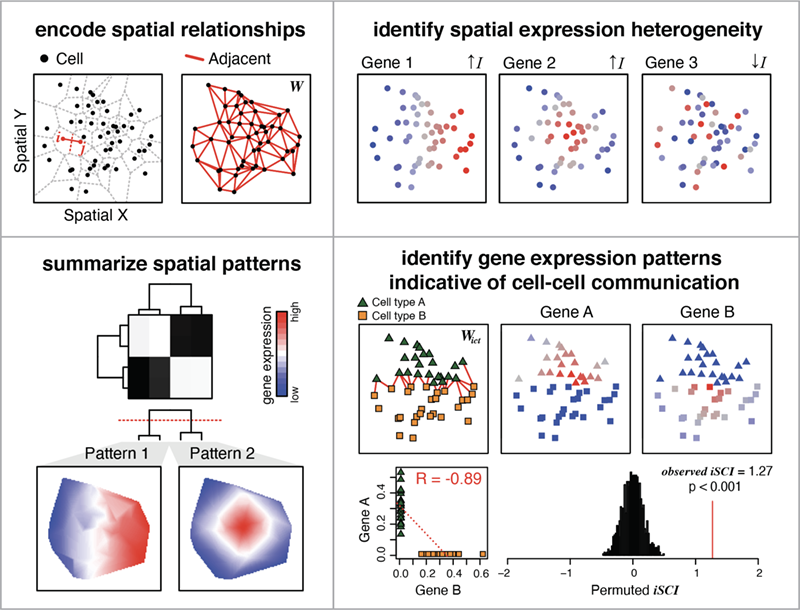
In a manner that:
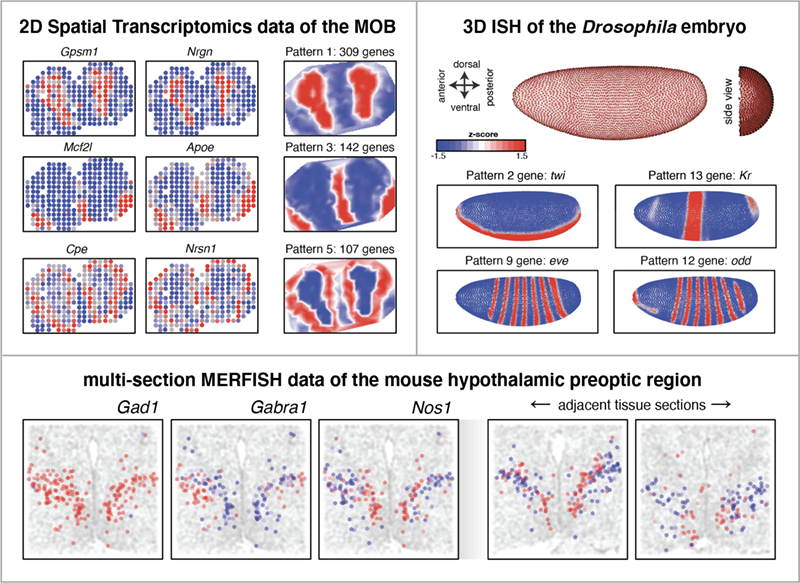
- Accomodates 2D, multi-section, and 3D spatial data
- Is robut to variations in cellular densities, distortions, or warping common to tissues
- Is highly scalable to enable analysis of 10,000s of genes and 1,000s of cells within minutes
- Is applicable to diverse spatial transcriptomics technologies
Installation
To install MERINGUE, we recommend using remotes:
# install.packages(remotes)
require(remotes)
remotes::install_github('JEFworks-Lab/MERingue', build_vignettes = TRUE)
Tutorials
-
Multi-section 3D Breast Cancer Spatial Transcriptomics Analysis
-
Understanding MERINGUE’s Spatial Cross-Correlation Statistic using Simulations
Contributing
We welcome any bug reports, enhancement requests, general questions, and other contributions. To submit a bug report or enhancement request, please use the MERINGUE GitHub issues tracker. For more substantial contributions, please fork this repo, push your changes to your fork, and submit a pull request with a good commit message.