Associations between cell localization and PC1 and 2 expression
Description
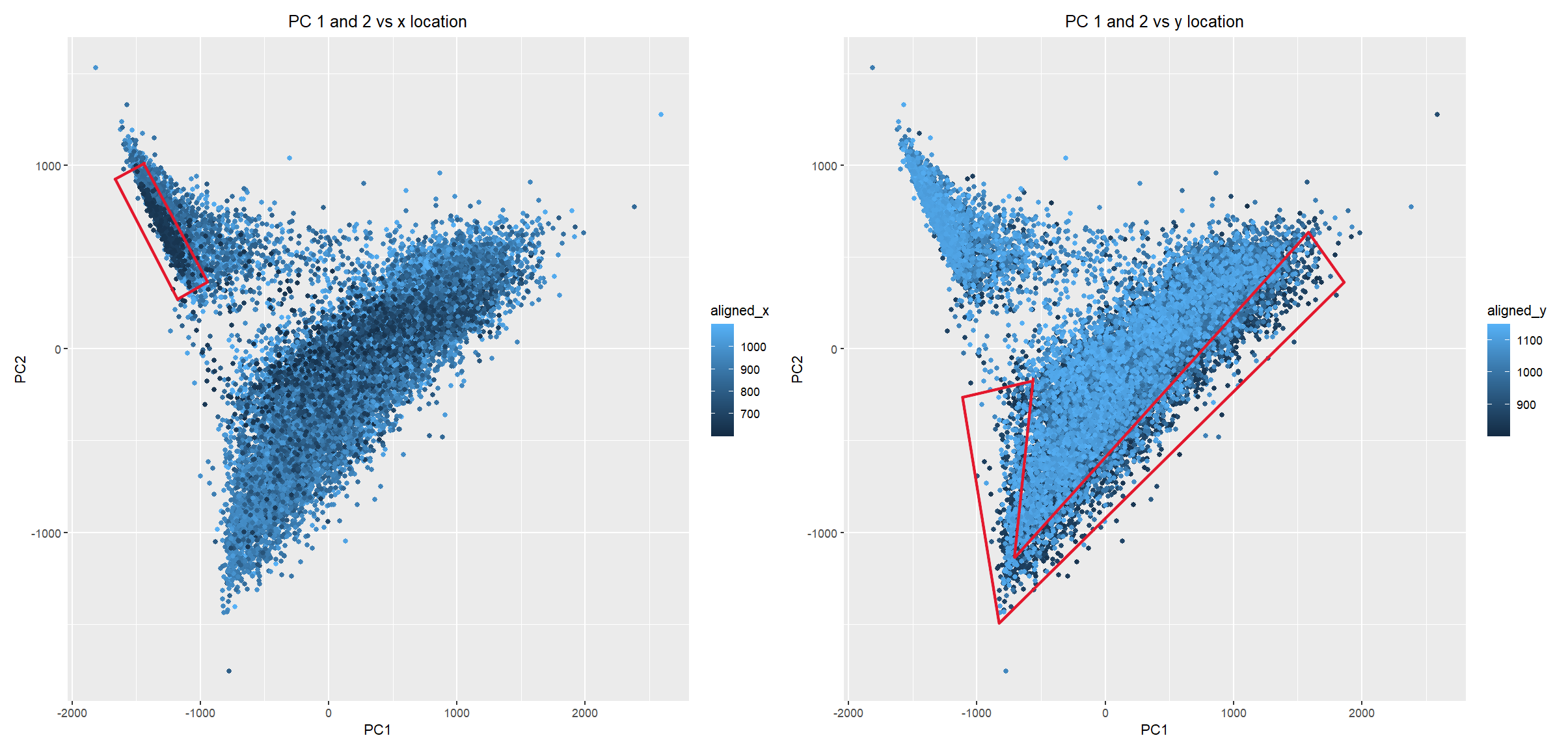
I have chosen to focus on how cells relate in the gene expression vs. physical space. I analyzed this by creating two visualizations- one which looks at the x alignment of the cells compared to the main two principle components, and one which looks at the y alignment of the cells. I used color as the main encoding that showed the x and y location, and used points as my geometric primitives to represent each cell. I also used the Gestalt principle of enclosure to highlight key areas on each of the visualizations. The enclosures represent areas of cells that are close to each other (similar colors), which also have relatively similar gene expression profiles. In the x-location visualization, this profile is correlated with high PC2 and low PC1 expression. However, in the y-location visualization, the enclosure shows that cells that are aligned close to the bottom of the image all appear to have low scores for both PC1 and PC2 expression.
Code
library(ggplot2)
library(patchwork)
file <- "data/pikachu.csv.gz"
data <- read.csv(file)
data[1:5,1:10]
##How do cells relate in gene expression space versus physical space?
#For example, are cells that are more transcriptionally similar to each other
#also physically closer to each other? minimum of two panels
pos <- data[, 5:6]
rownames(pos) <- data$cell_id
gexp <- data[, 7:ncol(data)]
rownames(gexp) <- data$barcode
names(pcs)
pcs$sdev
pcs$rotation[1:5,1:5] ## beta values representing PCs as linear combinations fo genes
pcs$x[1:5,1:5]
data$total_gene_expression <- rowSums(data[, 7:ncol(data)])
data$PC1 <- pcs$x[,1]
data$PC2 <- pcs$x[,2]
p1 <- ggplot(data) +
geom_point(aes(x = PC1, y=PC2, col=aligned_x)) +
ggtitle("PC 1 and 2 vs x location") +
theme(plot.title = element_text(hjust=0.5)) +
xlab("PC1") + ylab("PC2")
p2 <- ggplot(data) +
geom_point(aes(x = PC1, y=PC2, col=aligned_y)) +
ggtitle("PC 1 and 2 vs y location") +
theme(plot.title = element_text(hjust=0.5)) +
xlab("PC1") + ylab("PC2")
p1 + p2
