Question 4: Exploring the Effect of Varying Principal Components for Non-Linear Dimensionality Reduction
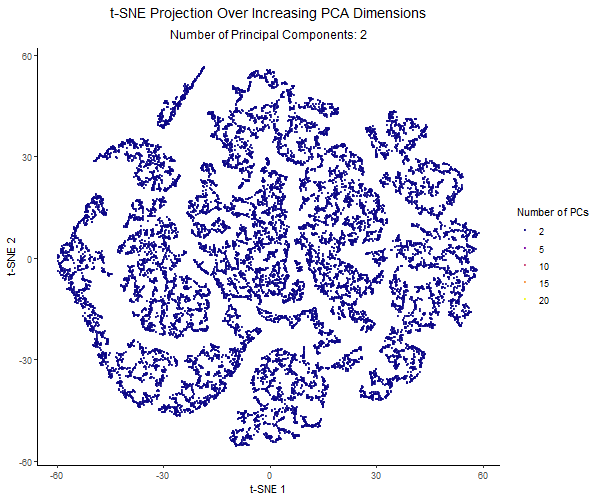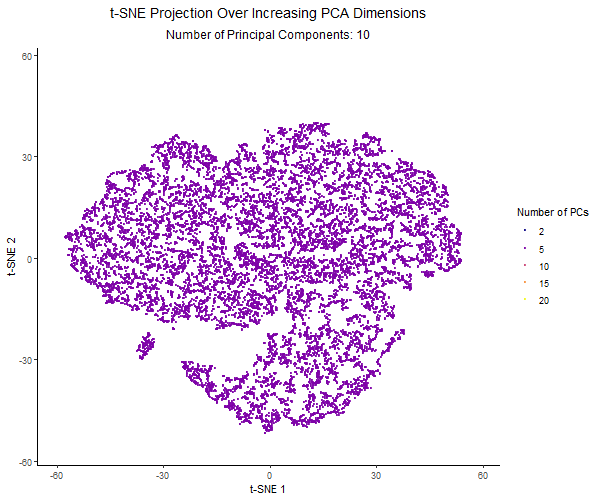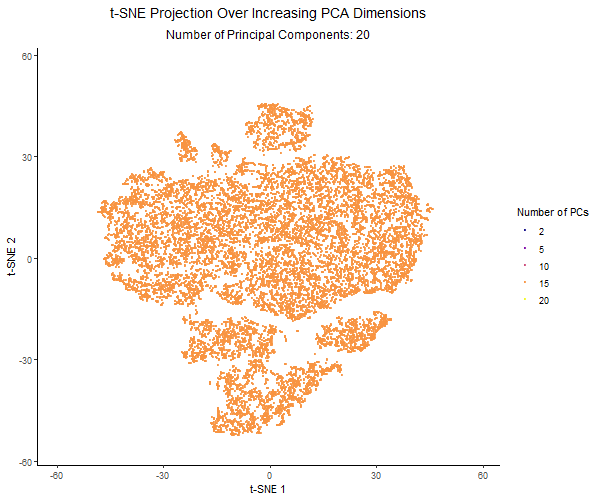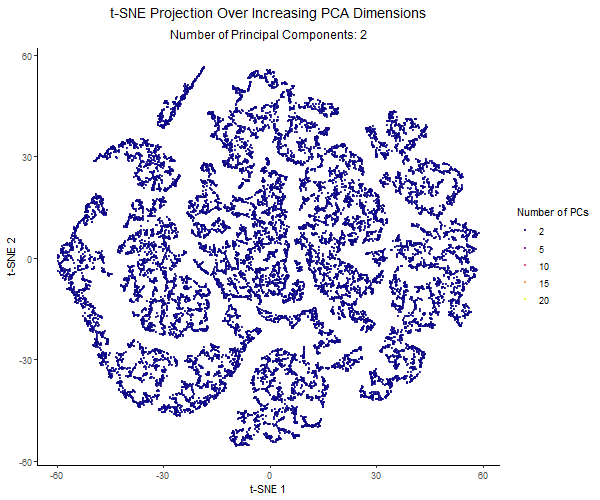
For this project, I created an animation using gganimate to visualize the effect of varying the number of principal components before applying t-SNE. I used the Pikachu dataset. First, I normalized the gene expression, and then ordered the genes by variance. I then applied PCA with a varying number of PCs (2, 5, 10, 15, 20). Increasing the number of PCs retains more of the dataset’s variance. Then I applied t-SNE to each PCA-reduced dataset and visualized the results across different PC counts using animation.
Each frame in the animation represents a different number of PCs, with the PC count dynamically displayed in the subtitle. The t-SNE embedding changes across frames, illustrating how the number of PCs used influences clustering and structure in the data. A dark-to-light color gradient represents the transition from low to high PC counts, making it easier to observe how the embeddings change.
From the animation, I noticed that when using only 2 PCs, the t-SNE plot is widely spread out, with a loosely connected structure and poorly separated clusters. As the number of PCs increased, the clusters became more distinct, and the overall structure of the data became clearer. However, with 20 PCs, the embeddings started to look noisier. The animation illustrates how the choice of number of PCs impacts the effectiveness of t-SNE for visualizing high-dimensional gene expression data. Using too few PCs can oversimplify the structure, while adding too many PCs can introduce unnecessary variance and distort the embedding.
# Load required libraries
library(gganimate)
library(ggplot2)
library(Rtsne)
library(dplyr)
library(patchwork)
library(here)
library(viridis)
# Load data
data <- read.csv('~/Desktop/pikachu.csv.gz', row.names = 1)
# Extract gene expression data
gexp <- data[, 6:ncol(data)]
# Normalize gene expression (Log Normalization)
gexpnorm <- log10(gexp / rowSums(gexp) * mean(rowSums(gexp)) + 1)
# Select top genes by variance
topgene <- names(sort(apply(gexpnorm, 2, var), decreasing = TRUE))
gexpfilter <- gexpnorm[, topgene]
# Perform PCA
pcs <- prcomp(gexpfilter)
# Define PCA components to use
pcs_list <- c(2, 5, 10, 15, 20)
# Run t-SNE for different PCA dimensions
tsne_results <- lapply(pcs_list, function(n) {
Rtsne(pcs$x[, 1:n], dims = 2, perplexity = 30)$Y
})
# Create a combined dataframe
df_list <- list()
for (i in seq_along(pcs_list)) {
df_tmp <- data.frame(
PC_X = pcs$x[,1], PC_Y = pcs$x[,2], # Keep original PC1, PC2
emb_x_1 = tsne_results[[i]][,1], emb_x_2 = tsne_results[[i]][,2],
gene = rowSums(gexpfilter), # Gene expression
order = pcs_list[i]
)
df_list[[i]] <- df_tmp
}
# Combine all into a single dataframe
df <- bind_rows(df_list)
# Set consistent axis limits across all frames
x_limits <- range(df$emb_x_1, na.rm = TRUE)
y_limits <- range(df$emb_x_2, na.rm = TRUE)
# Explicitly set factor levels to include ALL PCA values
df$order <- factor(df$order, levels = c(2, 5, 10, 15, 20))
# Create smooth animated plot using transition_time()
p <- ggplot(df, aes(x = emb_x_1, y = emb_x_2, color = order)) +
geom_point(size = 0.5, alpha = 0.8) +
scale_color_viridis_d(option = "plasma") +
theme_classic() +
xlim(x_limits) + ylim(y_limits) +
labs(
title = "t-SNE Projection Over Increasing PCA Dimensions",
subtitle = "Number of Principal Components: {closest_state}",
x = "t-SNE 1",
y = "t-SNE 2",
color = "Number of PCs"
) +
theme(
legend.position = "right",
plot.title = element_text(hjust = 0.5, size = 14, face = "bold"),
plot.subtitle = element_text(hjust = 0.5, size = 12)
) +
transition_states(order, transition_length = 2, state_length = 1) +
view_follow(fixed_x = TRUE, fixed_y = TRUE)
# Animation
animation <- animate(p, height = 500, width = 600, nframes = 400, fps = 30, renderer = gifski_renderer())
# Save animation
anim_save(here("kamiemueller.gif"), animation)
animation
