Spotting cytotoxic T-cells in Pikachu (imaging) dataset
Homework 1
1. What data types are you visualizing?
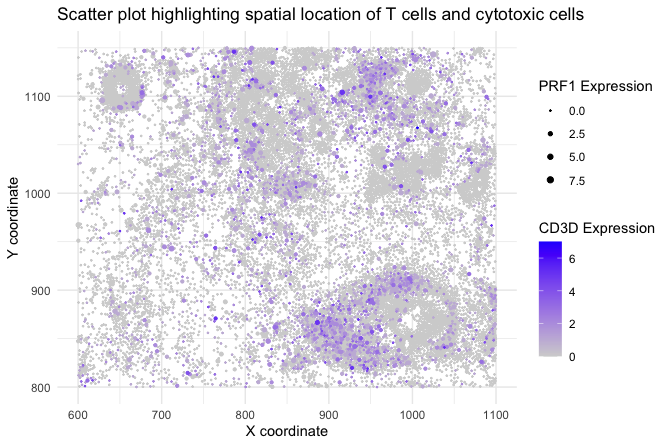
I am visualizing quantitative data of the expression of CD3D and PRF1 genes (2 quantitative variables) for each cell. Also, I am showing spatial data regarding the x,y centroid positions for each cell.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I am using the geometric primitive of points to represent each cell.
To encode expression counts of CD3D gene I am using the visual channel of color, and size for PRF1 gene.
For variables “aligned_x” and “aligned_y” I am using the visual channel of position along the x axis and y axis, respectively. That allows me to take advantage of the spatial location and compare it with any tissue staining I might have (e.g. H&E).
3. What about the data are you trying to make salient through this data visualization?
My data visualization seeks to make more salient the spatial location of T cells within the tissue slide, which are immune cells positive for CD3D expression. On top of that, I want to highlight cytotoxic cells bearing a high PRF1 expression.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
My plot is using the similarity principle. Cells that have a similar CD3D expression have a similar color. In the same way, all cells with high levels of PRF1 will be bigger than cells with lower expression levels.
5. Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
# Import libraries
library(ggplot2)
library(tidyverse)
# Read data
file <- '/Users/kmlanderos/Documents/Johns_Hopkins/Spring_2025/Genomic_Data_Visualization/genomic-data-visualization-2025/data/pikachu.csv.gz'
data <- read.csv(file, row.names=1)
# Number of cells
nrow(data)
# Number of genes
ncol(data[,6:ncol(data)])
# Create plot
data %>% arrange(CD3D) %>% mutate(cell_area=log2(cell_area), CD8_CD4_ratio=CD8A/CD4) %>%
ggplot(aes(x=aligned_x, y=aligned_y, col=CD3D)) +
scale_colour_gradient(low = 'lightgrey', high='#2810fe') +
#geom_point(size = 1) +
geom_point(aes(size = PRF1)) +
scale_size_continuous(range = c(0.2, 2)) +
theme_minimal() +
labs(
title = "Scatter plot highlighting spatial location of T cells and cytotoxic cells",
x = "X coordinate",
y = "Y coordinate",
size = "PRF1 Expression",
color = "CD3D Expression",
shape = "CD4+/CD8+ T cell"
)
