Spatial Distribution of SAMD11 Expression Levels
1. What data types are you visualizing?
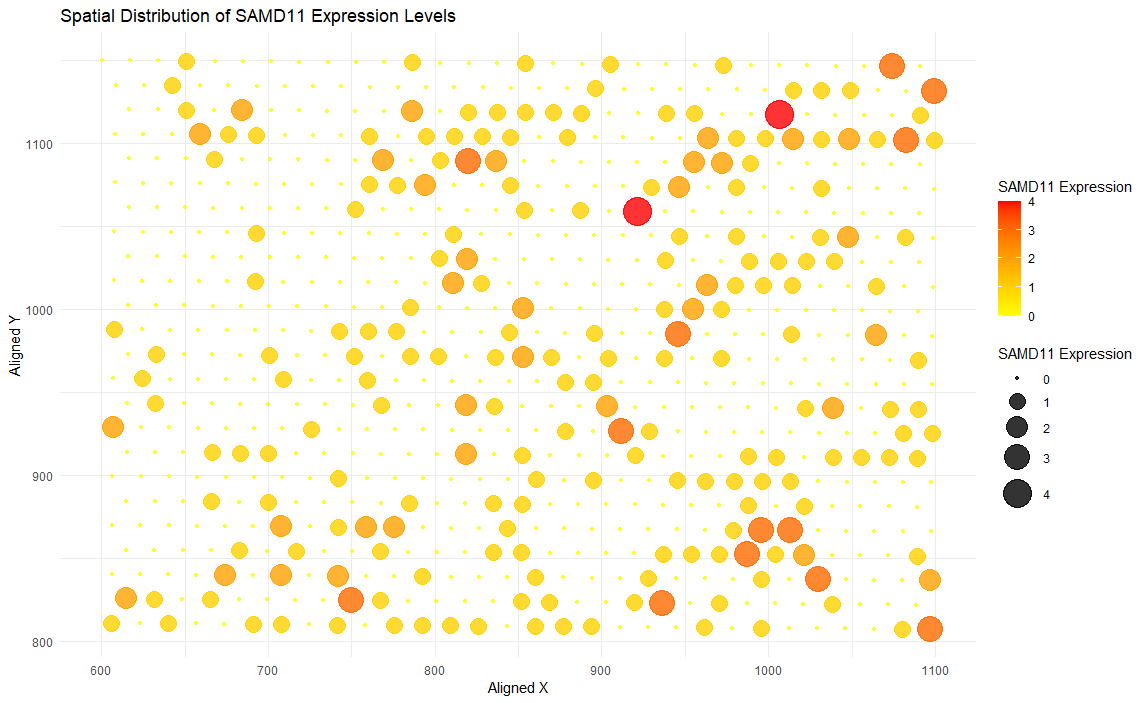
I am visualizing quantitative data representing SAMD11 gene expression levels, and spatial data regarding the x and y aligned positions of data points within the tissue sample.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I am using the geometric primitive of points to represent data points in the tissue sample. To encode the expression levels of SAMD11 gene, I am using various visual channels: Visual channel of location: I use it to encode the aligned x- and y- coordinate of each circle, representing the spatial location of each point. Visual channel of color: I establish a color gradient to encode SAMD11 expression levels. The color gradient ranges from yellow (low expression) to red (high expression). Visual channel of size: I use it to distinguish between expression levels: the radius of the circles is proportional to the magnitude of SAMD11 expression levels.
3. What about the data are you trying to make salient through this data visualization?
The data visualization tries to highlight the spatial distribution of SAMD11 gene expression levels within the tissue sample. As I map expression levels to circle size and color gradient and overlay those information on aligned spatial coordinates, clusters with varying SAMD11 gene expression levels could be determined (some regions, for example, have mostly orange or red dots, pointing to high SAMD11 expression).
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
I am using Gestalt principles of similarity and proximity in my data visualization. For similarity - data points with similar SAMD11 gene expression levels are represented with similar colors (used from the color gradient) and circle sizes, which lets the audience perceive that those data points are in a related group based on expression level. For proximity - Data points (circles) that are spatially close to each other will be perceived as a cluster based on spatial location, and this will help identify potential cluster patterns in gene expression.
5. Code (paste your code in between the ``` symbols)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
# read in dataset
file <- 'D:/genomic-data-visualization-2025/data/eevee.csv.gz'
data <- read.csv(file)
library(ggplot2)
# plot
ggplot(data, aes(x = aligned_x, y = aligned_y, color = SAMD11, size = SAMD11)) +
geom_point(alpha = 0.8) +
scale_color_gradient(low = "yellow", high = "red") +
scale_size_continuous(name = "SAMD11 Expression", range = c(1, 10)) + # adjust size scale
labs( # set labels
title = "Spatial Distribution of SAMD11 Expression Levels",
x = "Aligned X",
y = "Aligned Y",
color = "SAMD11 Expression"
) +
theme_minimal()
# Reference for ggplot
# https://rstudio.github.io/cheatsheets/data-visualization.pdf
