Spatial Scatter Plot of Number of Total Transcription
1. What data types are you visualizing?
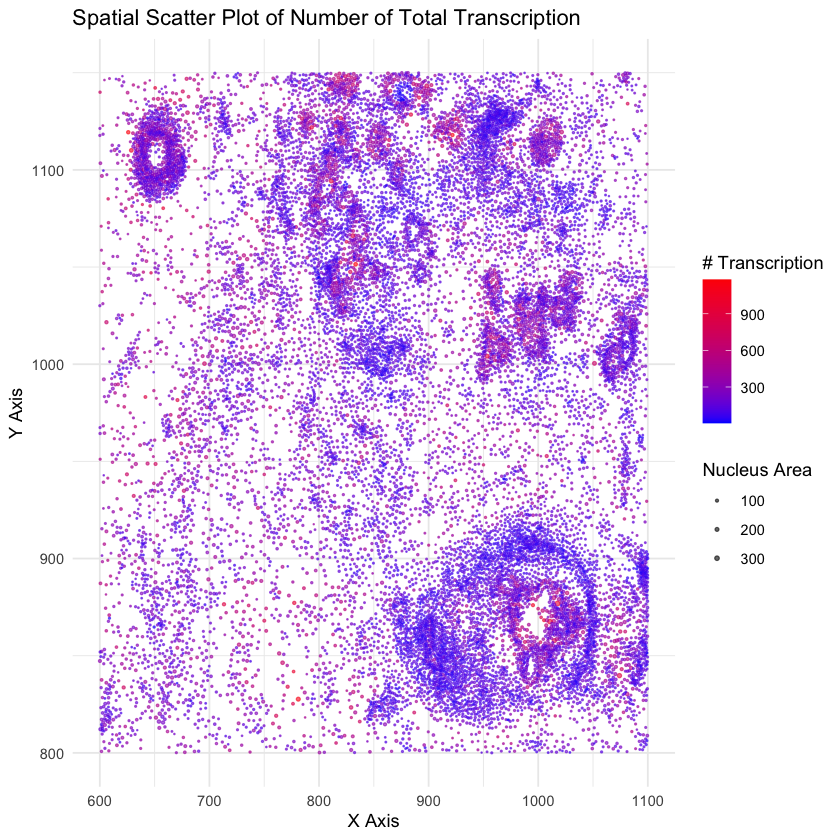
I am visualizing quantitative data of the number of expressed genes for each cell, quantitative data of the area for each cell, and spatial position of x,y to show the position of each cell.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I am using the area of the point to render each cell. I’m using the size of the points to represent the size of the area of the nucleus of the cell, and the color to show the number of expressed genes for that cell. To encode the position of the cell on the plate, I’m using the x y position of the circle in the image.
3. What about the data are you trying to make salient through this data visualization?
My data visualization seeks to make more salient the position (and distribution) of cells in the plate using the position in the figure.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
Proximity: items (cells) that are close together in 2D plane are considered grouped.
5. Code (paste your code in between the ``` symbols)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
file <- "/Users/sky2333/Downloads/genomic-data-visualization-2025/data/xenium.csv"
data <- read.csv(file)
data["sum"] <- rowSums(data[,7:ncol(data)])
ggplot(data, aes(x = aligned_x, y = aligned_y, color = sum, size = nucleus_area)) +
geom_point(alpha = 0.6) +
scale_color_gradient(low = "blue", high = "red") +
scale_size(range = c(0.01, 1)) +
theme_minimal() +
labs(
title = "Spatial Scatter Plot of Number of Total Transcription",
x= "X Axis",
y= "Y Axis",
color = "# Transcription",
size = "Nucleus Area"
)
