HW1 for Yi Yang
1. What data types are you visualizing?
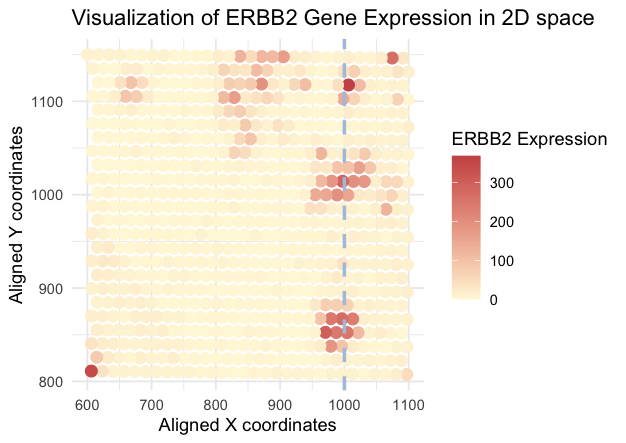
I’m trying to visualize the spatial (location of ERBB2 expression) and quantitative (ERBB2 expression) data.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
Geometric primitives includes points (expression data) and lines (specific x coordinate), while visual channels include color for quantitative expression data, position for spatial data.
3. What about the data are you trying to make salient through this data visualization?
I’m trying to make the spatial distribution of ERBB2 expression salient in 2D space, especially the enrichment around x = 1000.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
To accomplish this, color is used to highlight variation in ERBB2 expression, where close red points indicate the ERBB2 expression enrichment; the highlighted line draws the attention to the patterns of ERBB2 expression at x = 1000, which might correlate to the potential biological significance (similarity & proximity & enclosure).
5. Code (paste your code in between the ``` symbols)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
file <- '~/Desktop/genomic-data-visualization-2025/data/eevee.csv.gz'
data <- read.csv(file)
library(ggplot2)
ggplot(data, aes(x = aligned_x, y = aligned_y)) +
geom_point(aes(color = ERBB2), size = 3) +
scale_color_gradient(low = '#FFF8DC', high = "#CD5555") +
geom_vline(xintercept = 1000, linetype = "dashed", color = "#B0C4DE", size = 1) +
labs(
title = "Visualization of ERBB2 Gene Expression in 2D space",
x = "Aligned X coordinates",
y = "Aligned Y coordinates",
color = "ERBB2 Expression"
) +
theme_minimal()
