Homework 1 Submission
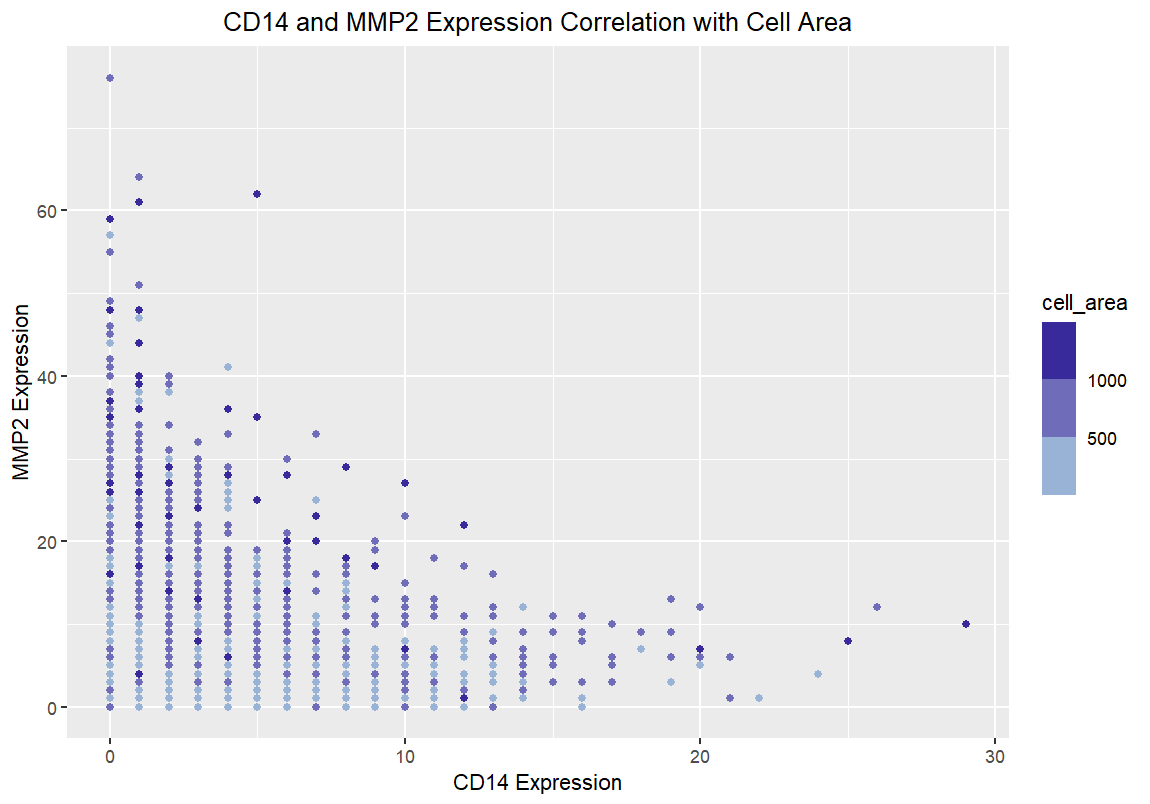
I am visualizing the gene expression of CD14 and MMP2 and how the expression of these genes relates to cell size.
What data types are you visualizing?
I am visualizing quantitative data in the form of gene expression profiles of CD14 and MMP2. I am also visualizing cell area as ordinal data. While cell area is technically quantitative, I chose to use bins to visualize cell area rather than a continuous color gradient. This set of bins puts the cell area into categories, but since area is still mathematically relevant, this makes it ordinal data.
What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
The only geometric primitives I am using are points, which are being used to display the cells that have CD14 and MMP2, as well as their respective expressions. In terms of channels, I am using the position of the points to show relative CD14 and MMP2 expressions, and I use the color of the points to represent the cell area.
What about the data are you trying to make salient through this data visualization?
I found a source [1] that claims that mostly macrophages express both CD14 and MMP2, and I know that macrophages are incredibly large cells. As such, I wanted to make it salient that the more CD14 and MMP2 are expressed together, the larger the cells will be (thus having a darker color).
- Cui, Q., Wang, X., Zhang, Y. et al. Macrophage-Derived MMP-9 and MMP-2 are Closely Related to the Rupture of the Fibrous Capsule of Hepatocellular Carcinoma Leading to Tumor Invasion. Biol Proced Online 25, 8 (2023). https://doi.org/10.1186/s12575-023-00196-0
What Gestalt principles and/or knowledge about perceptiveness of visual encodings are you using to accomplish this?
I know that saturation is a better encoding than color, and in particular, viewer’s eyes are drawn to higher saturations. As such, I made sure the higher cell area, which I want to draw attention to, was the more saturated color. I am also using the Gestalt principle of proximity, because the points which have very little expression of each gene are all quite close together, so they are perceived as related.
Code
library(ggplot2)
file <- "data/pikachu.csv.gz"
data <- read.csv(file)
#Code used to center the title (line 11) taken from
#https://www.geeksforgeeks.org/how-to-change-position-of-ggplot-title-in-r/
ggplot(data) +
geom_point(aes(x = CD14, y=MMP2, col=cell_area,)) +
scale_colour_steps(low = 'lightblue', high='darkblue') +
ggtitle("CD14 and MMP2 Expression Correlation with Cell Area") +
theme(plot.title = element_text(hjust=0.5)) +
xlab("CD14 Expression") + ylab("MMP2 Expression")
