Spatial Distribution and Correlation of ACTA2 and ACTC1 Expression
1. What data types are you visualizing?
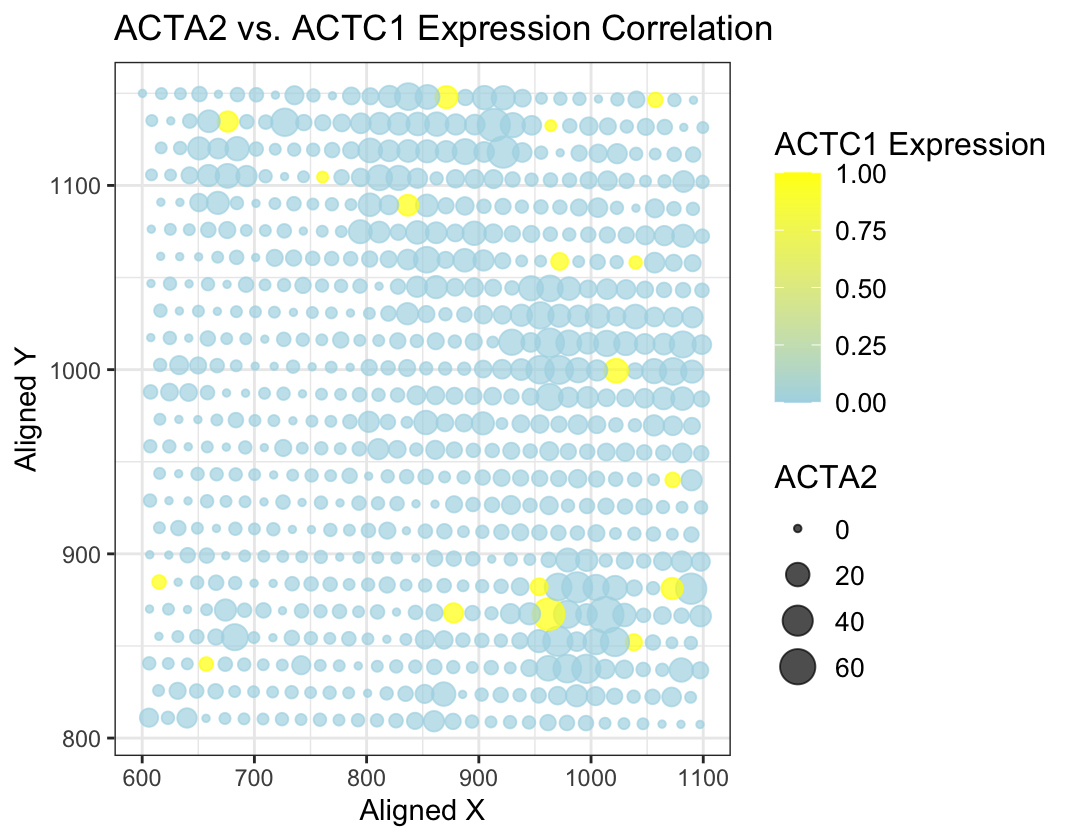
I am visualizing quantitative data of the expression correlation between ACTA1 and ACTA12 genes with its expression levels and spatial coordinates.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
The scatter plot is used as the geometric primitive, where each point’s position corresponds to aligned_x and aligned_y. The color of each point encodes ACTC1 expression levels, with a gradient from light blue to yellow, and the size of the points represents ACTA2 expression. This combination of encoding enables to visually assess the correlation between the two gene expressions and their spatial distribution.
3. What about the data are you trying to make salient through this data visualization?
The visualization emphasizes the correlation between ACTA2 and ACTC1 expressions by encoding their values through color and size. It also explores how these gene expressions vary in different spatial regions, indicated by aligned_x and aligned_y. This helps identify potential patterns or trends in gene expression relative to the spatial positioning.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
The proximity principle helps group data points that are spatially close together, making it easier to identify spatial patterns. The similarity of color and size aids in detecting correlations between ACTA2 and ACTC1 expressions.
5. Code (paste your code in between the ``` symbols)
1
2
3
4
5
6
7
8
9
10
11
file <- '~/Desktop/genomic-data-visualization-2025/data/eevee.csv.gz'
data <- read.csv(file)
data[1:100,1:100]
library(ggplot2)
ggplot(data) +
geom_point(aes(x = aligned_x, y = aligned_y, col = ACTC1, size = ACTA2), alpha = 0.7) +
scale_color_gradient(low = 'lightblue', high = 'yellow', name = "ACTC1 Expression") +
labs(x = "Aligned X", y = "Aligned Y", title = "ACTA2 vs. ACTC1 Expression Correlation") +
theme_bw() +
theme(legend.title = element_text(size = 12), legend.text = element_text(size = 10))
