Generation of Heatmap Expressing Top 20 Genes Within Pikachu Dataset
1. What data types are you visualizing?
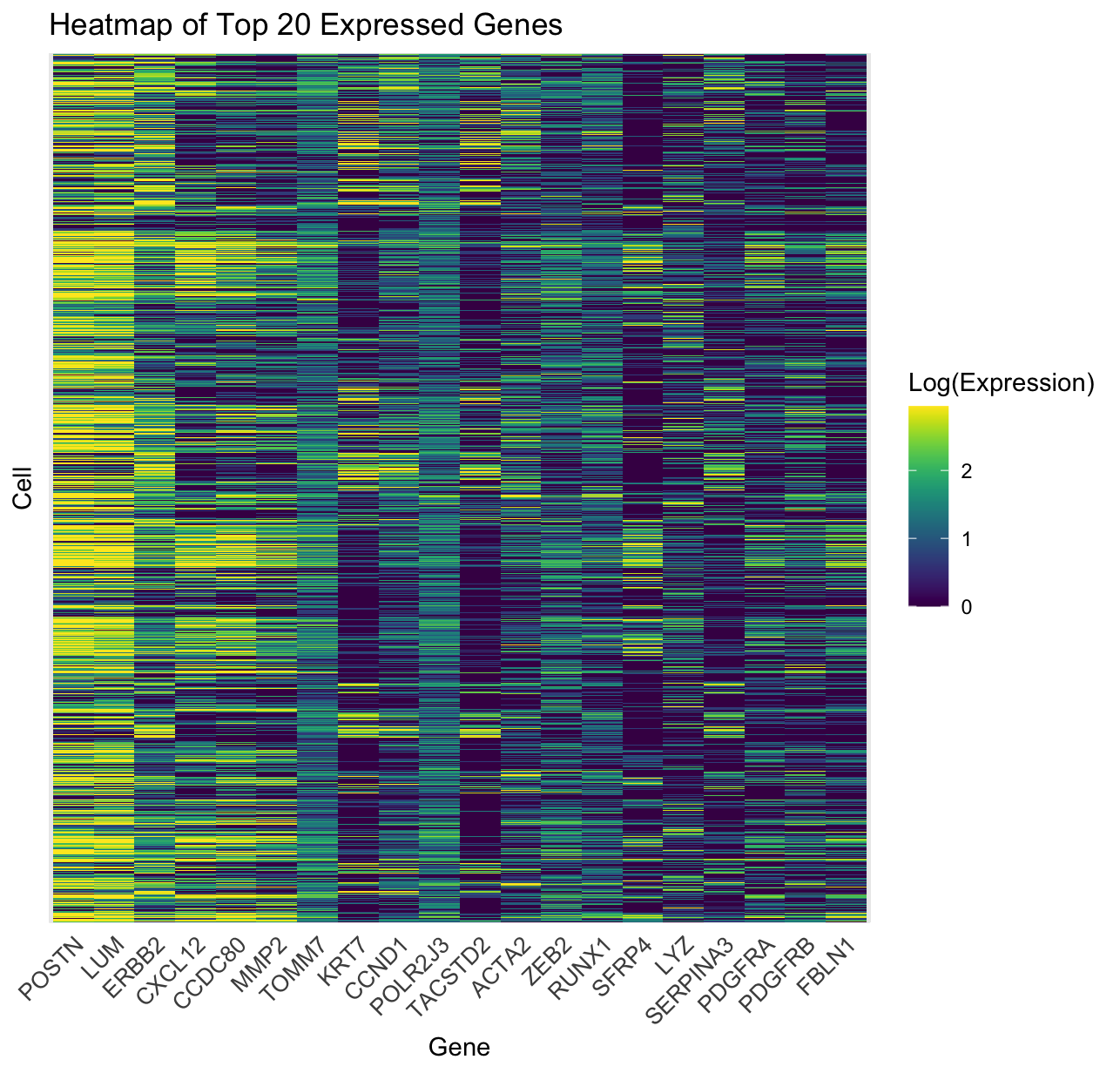
Within the Pikachu dataset that was visualized, gene expression levels across multiple individual cells proved to be a key mechanism in exploring spatial and comparative expression types. With this being said, I was able to visualize data types such as categorical data with gene names (present along the X-axis) and cell identification (present along the Y-axis), quantitative data with the expression level account of the top 20 genes for each cell within the dataset, as well as a form of ordinal data through the use of the log(Expression) scale to place all expression levels within a visual, ordinal scale.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
In order to visualize these data types, several data encodings were utilized. To express genetic expression level, a heatmap was generated with geometric primitives such as lines and areas being used through the geom_tile function in ggplot2. These geometric primitives utilize lines and rectangular areas to express the varying expression cells between the cells of each gene. In addition, the visual channel that was most significantly used was color through the viridis color map in order to depict the variation in gene expression levels across the dataset. The color hue channel demonstrated variation from low expression levels (purple color) to middle expression (green) to high expression (yellow color), allowing for expression changes to be clearly visualized. In addition, to improve the readability and visualization of the plot, limits of 10% and 90% quartiles were placed to eliminate outliers and allow for the heatmap to portray a color saturation channel that decreased the initially faint portrayal of the data.
3. What about the data are you trying to make salient through this data visualization?
Through the data visualization of this heatmap, the dataset is represented through genes expressed across cells. As a result, the data becomes more salient with clusters of high-expression genes being identified. Through patterns & clustering of the data with the use of visual color channels, cells with similar expression profiles appear as blocks of similar colors while also allowing for the easy visual comparison of which genes are co-expressed in multiple cells. In terms of visual differentiation of the data, the geometric primitives (lines and area) and visual property (color gradient) are differentiated, allowing for the immediate recognition of clusters of high expression without having to scan each line. The final attempt to enhance salience within the data is through the use of genes with the highest differential expression being encoded through both position (X-axis: gene names, Y-axis: cells) and color intensity, ensuring that the biologically significant trends within the heatmap are immediately salient.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
The Gestalt principles of visual perceptions that are being utilized in this visualization include proximity, similarity, and continuity. Proximity is demonstrated through the spatial arrangement of genes and cells being grouped within proximal groups and patterns while similarity is depicted through continual color gradients that group similar visual colors to demonstrate similar expression levels, allowing for the easy detection of trends within the data. Finally, the use of continuity allows for differences in expression levels to be smoothly present throughout the dataset while avoiding abrupt gaps and noise within the dataset.
5. Code (paste your code in between the ``` symbols)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
# Necessary libraries loaded
library(ggplot2)
library(data.table)
library(reshape2)
library(scales) # For log transformation
# Loads Pikachu Dataset
file_path <- "~/Desktop/genomic-data-visualization-2025/data/pikachu.csv.gz"
data <- fread(file_path)
# Initial 6 columns removed to solely include gene dataset (V1, cell_ID, cell_area, nucleus_area, aligned_x, aligned_y)
gene_data <- data[, -(1:6), with = FALSE]
# Adds Cell ID column (assuming each row is a unique cell)
gene_data[, Cell_ID := .I] # Creates a unique cell index
# Finds the top 20 most expressed genes out of gene dataset
top_genes <- names(sort(colSums(gene_data[, -c("Cell_ID"), with = FALSE], na.rm = TRUE), decreasing = TRUE))[1:20]
# Creates a subset of the dataset to only include the top 20 genes + Cell_ID
heatmap_data <- gene_data[, c("Cell_ID", top_genes), with = FALSE]
# Converts data from wide format to long format for ggplot use
heatmap_melt <- melt(heatmap_data, id.vars = "Cell_ID")
# Renames columns to Cell, Gene, and Expression
colnames(heatmap_melt) <- c("Cell", "Gene", "Expression")
# Applies log transformations for better contrast within the colors of the heatmap
heatmap_melt$Expression <- log1p(heatmap_melt$Expression) # log(x + 1) to prevent log(0)
# Calculates and generates a stronger color contrast using percentiles for the colors of the heatmap
p95 <- quantile(heatmap_melt$Expression, 0.95, na.rm = TRUE) # 95th percentile
p5 <- quantile(heatmap_melt$Expression, 0.05, na.rm = TRUE) # 5th percentile
# Creates final heatmap using viridis color map
ggplot(heatmap_melt, aes(x = Gene, y = as.factor(Cell), fill = Expression)) +
geom_tile() +
scale_fill_gradientn(colors = viridis::viridis(100), limits = c(p5, p95), oob = scales::squish) +
theme_minimal() +
labs(title = "Heatmap of Top 20 Expressed Genes", x = "Gene", y = "Cell", fill = "Log(Expression)") +
theme(
axis.text.x = element_text(angle = 45, hjust = 1, size = 10), # Rotate x-axis labels
axis.text.y = element_blank(), # Hide y-axis labels to reduce clutter
axis.ticks.y = element_blank(), # Remove y-axis ticks
legend.position = "right"
)
# Sources:
# https://monashbioinformaticsplatform.github.io/2015-09-28-rbioinformatics-intro-r/01-supp-addressing-data.html
# https://www.rdocumentation.org/packages/base/versions/3.6.2/topics/colSums
# https://dk81.github.io/dkmathstats_site/ranalysis-meltFunction.html
# https://www.geeksforgeeks.org/performing-logarithmic-computations-in-r-programming-log-log10-log1p-and-log2-functions/
# https://www.geeksforgeeks.org/create-quantiles-of-a-data-set-in-r-programming-quantile-function/
# https://r-charts.com/correlation/heat-map-ggplot2/
# https://cran.r-project.org/web/packages/viridis/vignettes/intro-to-viridis.html
# https://ggplot2.tidyverse.org/reference/theme.html
