Visualizing SDF4 and ERBB2 Expression
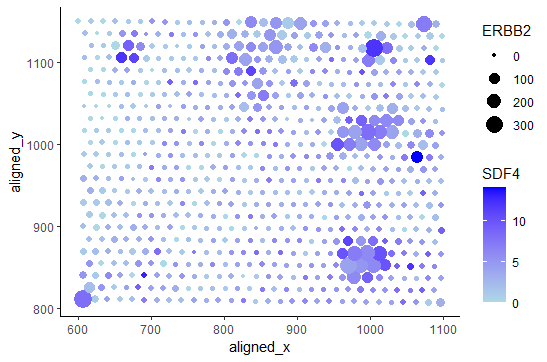
This visualization focuses on displaying SDF4 and ERBB2 Expression.
1. What data types are you visualizing?
I am visualizing quantitative data, specifically levels of SDF4 and ERBB2 expression along with their spatial data (where they are located in the x and y directions).
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I am using the geometric primitive of points with a visual channel of size (levels of ERBB2 expression) and color, specifically variations of the blue hue (levels of SDF4 expression). Position in space is also showed to represent location.
3. What about the data are you trying to make salient through this data visualization?
Through the visualization I am trying to make salient the amounts of expression of SDF4 (through color hue) and ERBB2 (through size). This particular visualization provides the ability to see both the levels of expression of SDF4 and ERBB2 seperately, as well as together. However, this visualization is intended to most easily display the relation between the expression of both of those genes. Additionally, this visualization makes salient the locations of the expressed genes.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
In this visualization I am using the Gestault principle of similarity. For example, different hues of the same color are used to express different amounts of expression of the same gene (SDF4). Points with the same size are used to express the same amount of expression of the ERBB2 gene.
5. Code:
1
2
3
4
5
6
7
8
file <- 'C:\\Hopkins School Stuff\\GenomicDataVis\\data\\eevee.csv.gz'
data <- read.csv(file)
library(ggplot2)
ggplot(data) + geom_point(aes(x = aligned_x, y = aligned_y,
col = SDF4 ,size = ERBB2)) +
scale_color_gradient(low ='lightblue', high = 'blue') +
theme_classic()
