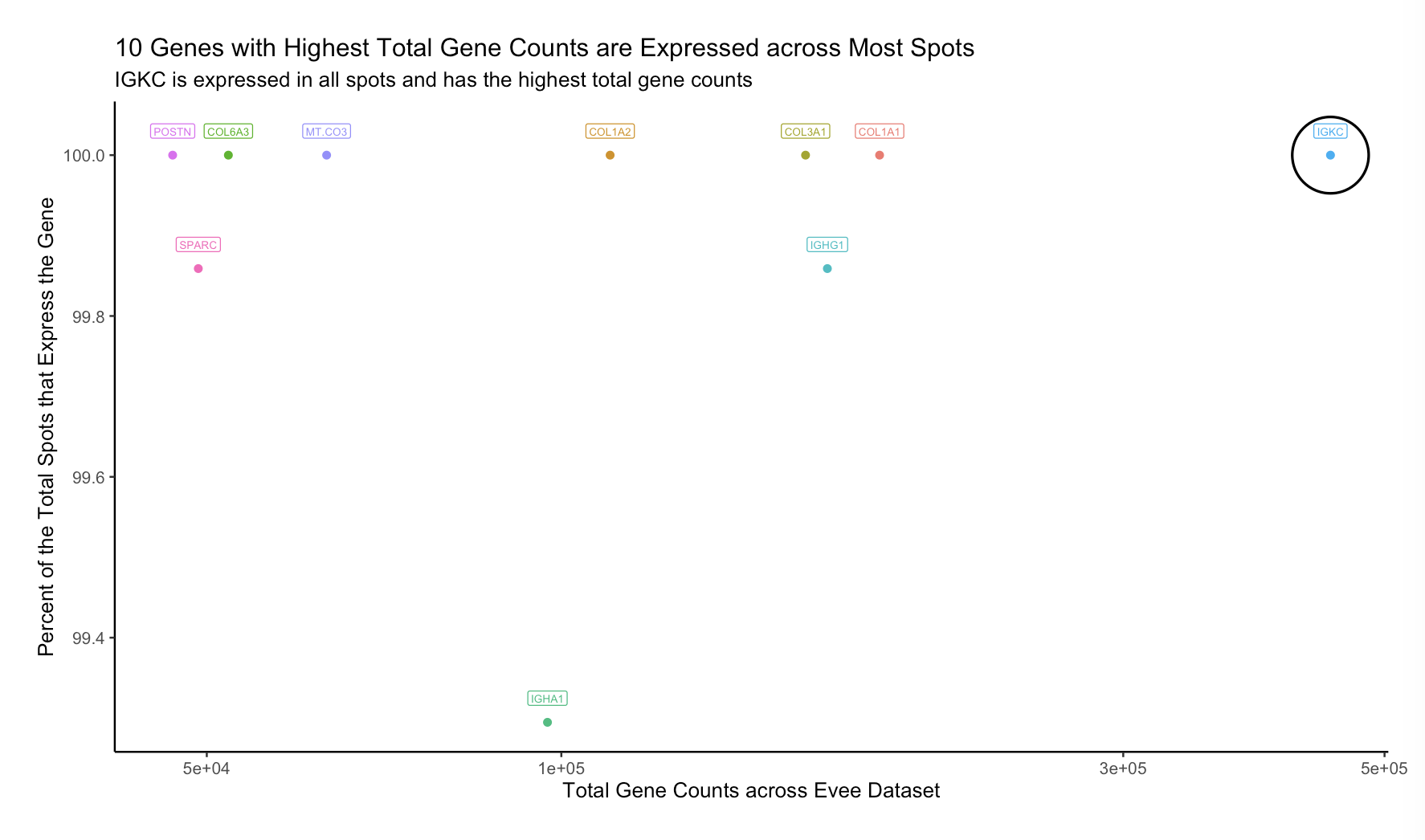
10 Genes with the Highest Counts are Expressed Across Most Spots
1. What data types are you visualizing?
For this data visualization of the Eevee spatial transcriptomic data, I visualized both categorical data, the 10 genes of interest, and two types of quantitative data, the total counts for each gene and the percent of the total spots that expressed each gene.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I used the geometric primitive of a point in my scatter plot, and I encoded the quantitative data of total counts for each gene using the x position, the quantitative data of the percent of the total spots that expressed each gene using the y position, and the categorical data of the gene names using color hue.
3. What about the data are you trying to make salient through this data visualization?
I am trying to make salient that all 10 of these highly expressed genes are expressed in a very high percentage of the total spots but also allow for easy comparison between these different genes. Also, I am particularly trying to make salient the fact that IGKC has both the highest total expression and non-zero expression in all spots.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
To accomplish these saliency goals, I used the Gestalt principles of similarity and enclosure and the knowledge of perceptiveness of visual encodings for quantitative and categorical data. I employed the Gestalt principle of similarity when choosing to make all data points and their corresponding label different colors to highlight that they denote different genes. The enclosure principle inspired my use of a circle to highlight IGKC in particular, though it is not a group and thus does not entirely fall under the Gestalt principle, because I know from this principle that the eye is drawn to enclosed points, and I wanted to attract attention to IGKC. Finally, position is known to be the best visual encoding for the perception of quantitative data, and hue is known to be the second best visual encoding for categorical information (after position, which I already employed), so I chose these encodings to allow an audience to best perceive my data.
5. Code (paste your code in between the ``` symbols)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
file <- '/Users/alexgorham/Desktop/genomic-data-visualization-2025/data/eevee.csv.gz'
data <- read.csv(file)
pos <- data[,3:4]
rownames(pos) <- data$barcode
gexp <- data[,5:ncol(data)]
rownames(gexp) <- data$barcode
summed_cols <- sapply(1:ncol(gexp), function(i) {
sum(gexp[,i])
})
sum_ordered_cols <- summed_cols[order(summed_cols, decreasing = TRUE)]
sum_ordered_gene_names <- colnames(gexp)[order(summed_cols, decreasing = TRUE)]
top_10_percent_cells_exp <- sapply(1:10, function(i){
(sum(gexp[,sum_ordered_gene_names[i]] != 0)/nrow(gexp))*100
})
x <- sum_ordered_cols[1:10]
y <- top_10_percent_cells_exp
df <- data.frame(x = sum_ordered_cols[1:10],
y = top_10_percent_cells_exp,
name = sum_ordered_gene_names[1:10])
library(ggplot2)
ggplot(df) +
geom_point(aes(x=x, y=y, colour = name)) +
theme_classic() +
geom_label(aes(x = x, y = y, label = name, colour = name), nudge_y = 0.03, size = 2) +
scale_x_log10() +
theme(legend.position = "none", plot.margin = margin(20, 20, 20, 20)) +
labs(
x = "Total Gene Counts across Evee Dataset",
y = "Percent of the Total Spots that Express the Gene",
title = "10 Genes with Highest Total Gene Counts are Expressed across Most Spots",
subtitle = "IGKC is expressed in all spots and has the highest total gene counts") +
geom_point(data = df[1, ], aes(x = x, y = y), shape = 1, size = 18, color = "black", stroke = 1)
