Comparison of Spatial Gene Expression of ESR1 and PGR
1. What data types are you visualizing?
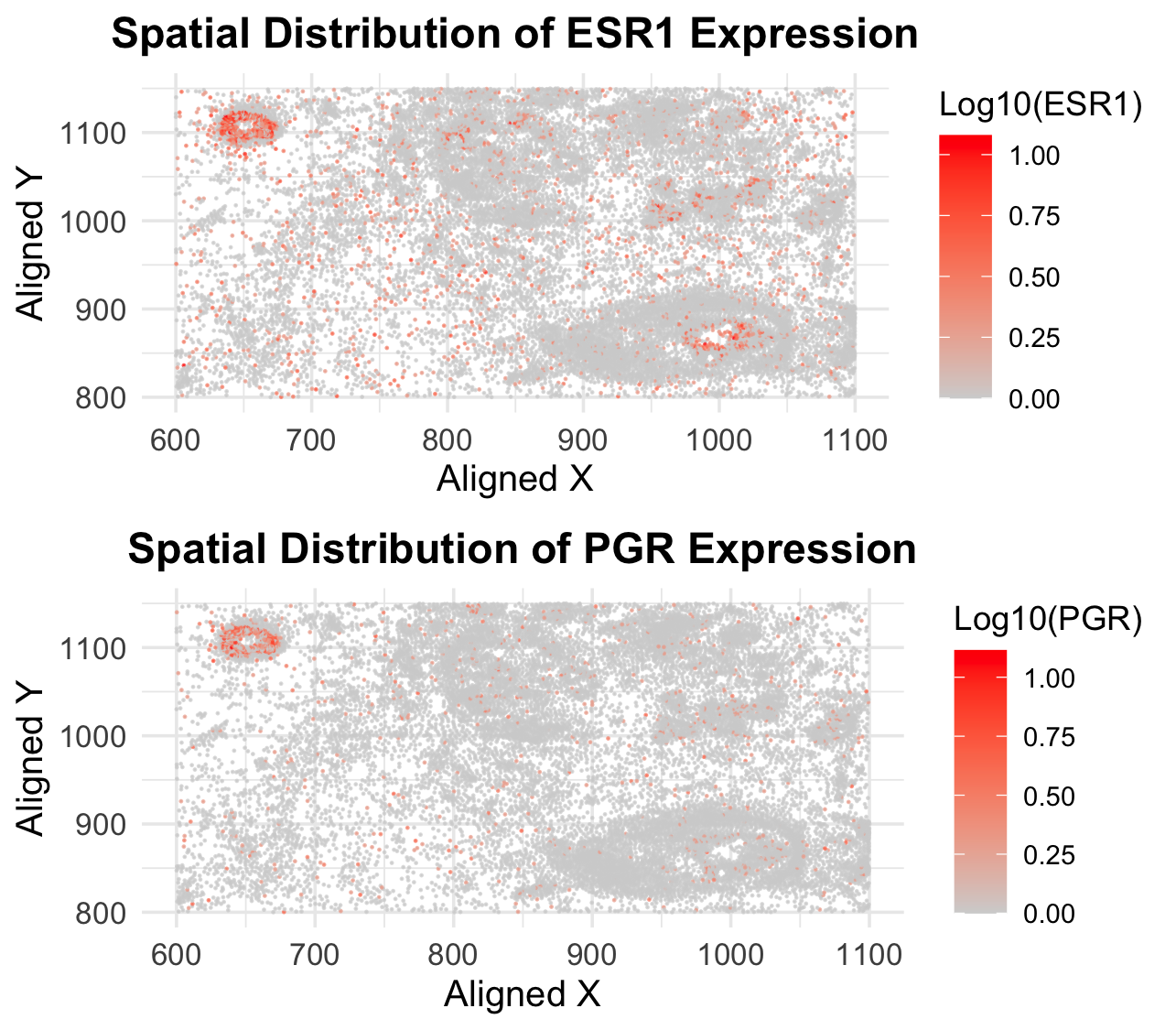
I am visualizing quantitative data representing the expression levels of the ESR1 and PGR genes for each cell. Additionally, I am visualizing spatial data that describes the x and y centroid positions of each cell within the tissue.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
Geometric Primitives: I am using points to represent each cell in the spatial context. Visual Channels: Color Hue and Saturation - The expression levels of the ESR1 and PGR genes are encoded using a color gradient. The gradient ranges from light grey (low expression) to bright red (high expression). This helps visually distinguish cells with varying levels of gene expression. Position - The spatial x and y positions of each cell are encoded using the x-axis and y-axis, respectively, to represent their physical locations within the tissue.
3. What about the data are you trying to make salient through this data visualization?
The visualization aims to highlight the spatial distribution of ESR1 and PGR gene expression across the tissue. By mapping gene expression levels to color and overlaying them on the spatial coordinates, I seek to reveal patterns or clusters of cells with similar expression profiles. This can help identify regions of the tissue where these genes are highly or lowly expressed, providing insights into their potential roles in cellular function or disease.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
Principle of Similarity: Cells with similar gene expression levels are encoded using similar colors (light grey to red), making it easy to group and compare them visually. Principle of Proximity: Cells that are close to each other in space are perceived as belonging to the same region or cluster, helping to identify spatial patterns in gene expression. Principle of Continuity: The smooth color gradient (light grey to red) creates a continuous visual flow, allowing viewers to intuitively interpret changes in gene expression levels.
5. Code (paste your code in between the ``` symbols)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
# Read file
data <- read.csv('~/Desktop/genomic_data_visualization/genomic-data-visualization-2025/data/pikachu.csv.gz', row.names=1)
# Load necessary libraries
library(ggplot2)
library(gridExtra)
# Apply log transformation to ESR1 and PGR
data$ESR1_log <- log10(data$ESR1 + 1)
data$PGR_log <- log10(data$PGR + 1)
breaks_labels <- seq(0, 1, by = 0.25)
# Plot for ESR1
p1 <- ggplot(data) +
geom_point(aes(x = aligned_x, y = aligned_y, col = ESR1_log), size = 0.01, alpha = 0.7) +
scale_colour_gradient(low = 'lightgrey', high = 'red', name = "Log10(ESR1)", breaks = breaks_labels) +
labs(
title = "Spatial Distribution of ESR1 Expression",
x = "Aligned X",
y = "Aligned Y"
) +
theme_minimal() +
theme(
plot.title = element_text(size = 14, face = "bold", hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
)
# Plot for PGR
p2 <- ggplot(data) +
geom_point(aes(x = aligned_x, y = aligned_y, col = PGR_log), size = 0.01, alpha = 0.7) +
scale_colour_gradient(low = 'lightgrey', high = 'red', name = "Log10(PGR)", breaks = breaks_labels) +
labs(
title = "Spatial Distribution of PGR Expression",
x = "Aligned X",
y = "Aligned Y"
) +
theme_minimal() +
theme(
plot.title = element_text(size = 14, face = "bold", hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
)
# Arrange plots
grid.arrange(p1, p2)
# Reference: https://rstudio.github.io/cheatsheets/html/data-visualization.html for ggplot codes
