Spatial Visualization of POSTN Expression in Tissue
What data types are you visualizing?
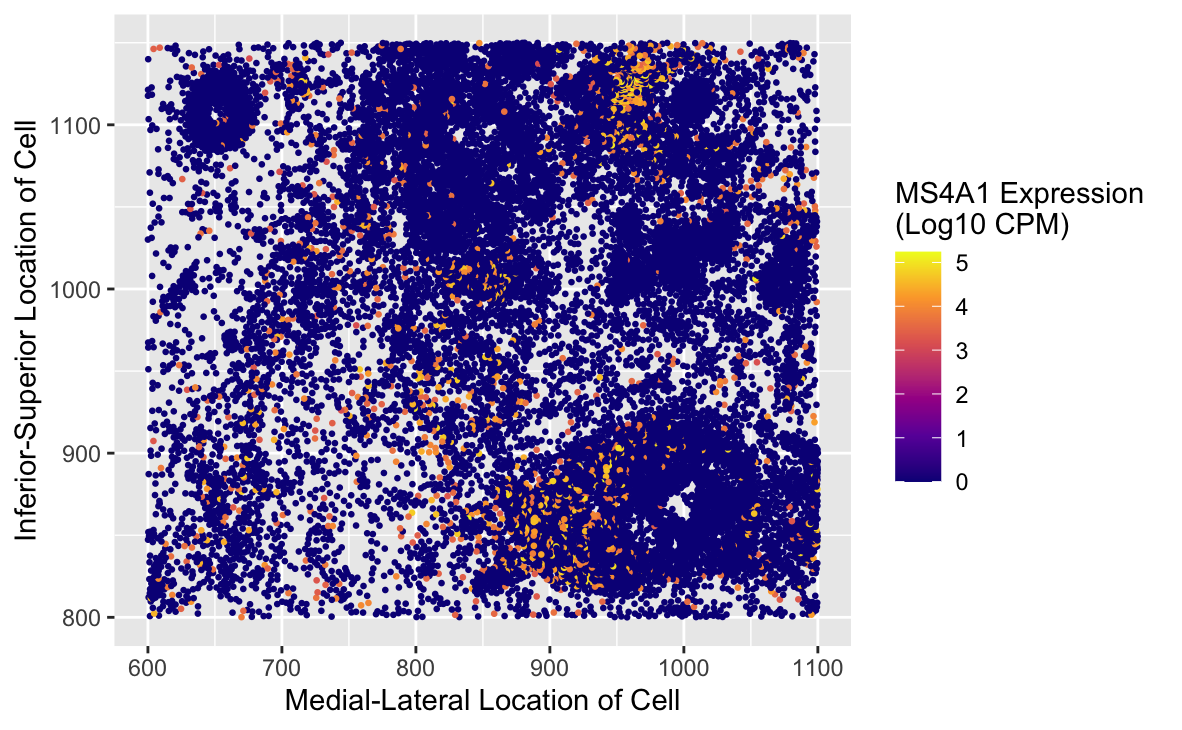
The data visualized represents the spatial distribution of MS4A1 expression. This is a gene encoding CD20, a well-known surface marker expressed on B-cells, which are a critical part of the adaptive immune system. The dataset originates from a spatial transcriptomics experiment and includes both spatial coordinates of cells and their respective gene expression levels. The data has been normalized using Counts Per Million (CPM) and transformed with log10(x + 1) scaling to account for cells with no measurable expression. Expression values range from 0 (no expression) to 5 (high expression) in the log10 scale.
What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
To represent individual cells in the tissue, I used points as geometric primitives. Each point represents a single cell. The following visual channels encode the data:
Position (x, y): These encode the spatial location of each cell within the tissue.
Color (hue): This encodes the expression level of MS4A1 on a per-cell basis. A gradient is applied, where higher expression levels are shown in warm colors (yellow, orange, red), and cells without measurable expression are blue.
This encoding leverages pre-attentive processing principles, as viewers can easily identify and differentiate high-expression cells by their warm hues.
What about the data are you trying to make salient through this data visualization?
The visualization highlights a key biological trend: MS4A1 expression (and by extension, B-cells) is localized to a specific region, suggesting a potential lymphoid aggregation. Such clusters may indicate immune activity, often associated with tissue damage, inflammation, or tumor microenvironments.
What Gestalt principles and/or knowledge about perceptiveness of visual encodings are you using to accomplish this?
To make this trend salient, I applied these Gestalt principles:
-
Similarity: Cells are colored based on expression levels, with warm colors representing B-cells and blue representing other cells. This differentiation emphasizes the grouping of B-cells.
-
Proximity: The spatial distribution of points naturally reveals areas where cells are clustered, reinforcing the localized nature of MS4A1 expression.
These principles guide viewers to perceive the pattern of B-cell clustering and their spatial relationship to other cells in the tissue.
#install.packages('viridis') #color gradients
library(viridis)
library(tidyverse)
library(ggplot2)
# Reading the data
data <- read.csv('~/Desktop/gdv_hw1/pikachu.csv.gz')
# Selecting the gene expression columns and the position columns
pos <- data[, c('aligned_x', 'aligned_y')]
rownames(pos) <- data[,1]
gexp <- data[, 7:ncol(data)]
rownames(gexp) <- data[,1]
# Counts per Million CPM normalization
numgenes <- rowSums(gexp)
normgexp <- gexp/numgenes*1e6
normgexp = log10(normgexp+1)
# Choose gene g
g <- "MS4A1"
# Create dataframe
df <- data.frame(x=pos[,1], y=pos[,2], gene = normgexp[,g])
# Plot dataframe
ggplot(df,aes(x=x, y=y)) +
geom_point(mapping = aes(col=gene),size=0.5) +
labs(x = "Medial-Lateral Location of Cell", y = "Inferior-Superior Location of Cell",
color = paste(g, "Expression \n(Log10 CPM)")) +
scale_color_viridis_c(option = "plasma")
