Spatial Distribution of Total Genes Expressed
1. What data types are you visualizing?
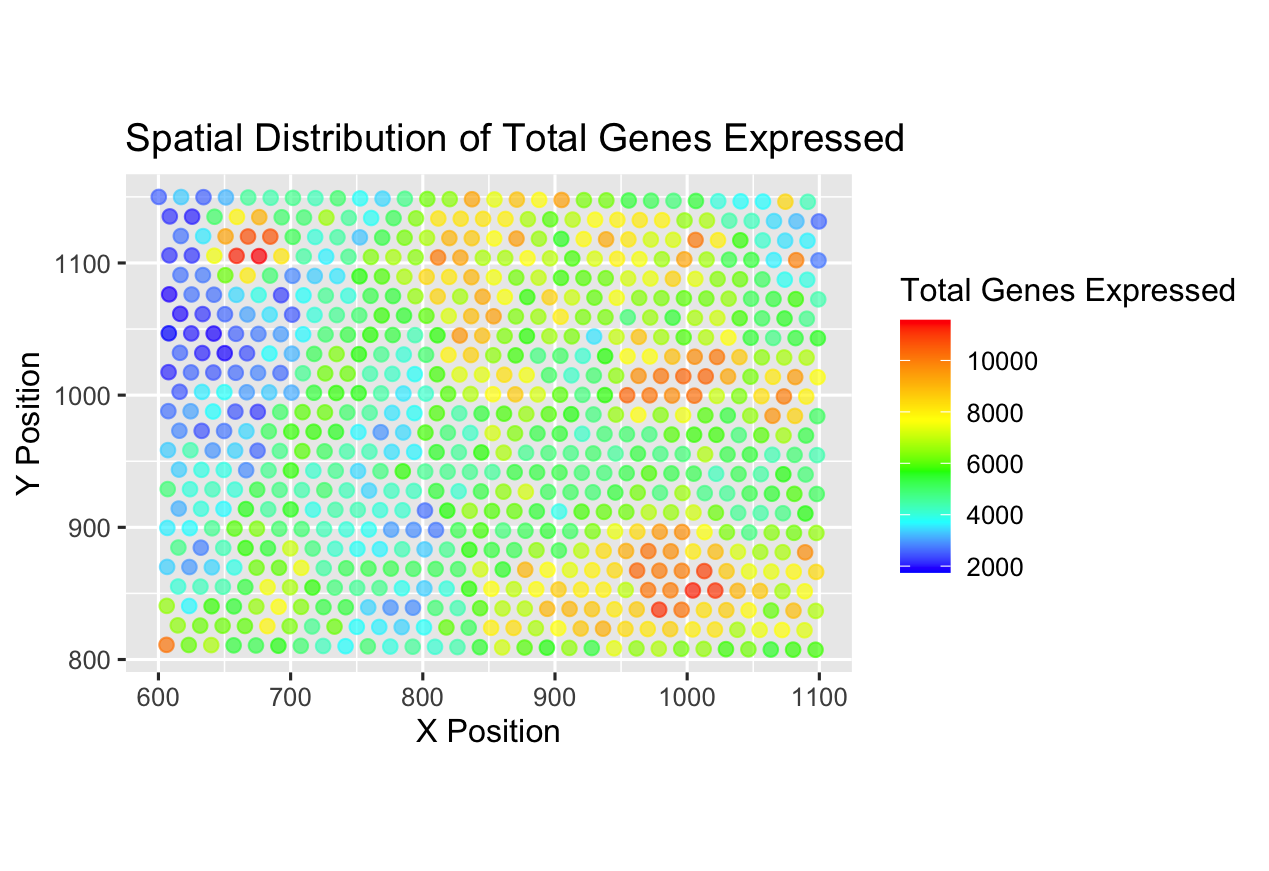
I am visualizing quantitative data of the total number of distinct genes expressed per spot. Additionally, I am visualizing spatial data for the aligned x- and y-coordinates that provide information regarding the physical location of the spot.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I am using the geometric primitive of points to represent each spot. For these points, I am using the visual channel of position to encode the location of each spot in the X-Y coordinate plane. To encode for the total number of genes expressed, I am using the visual channel of color and hue, allowing for visualization of the range of total number of genes present at a given spot.
3. What about the data are you trying to make salient through this data visualization?
My data visualization seeks to make more salient how the total number of genes expressed varies across different locations. Essentially, I am seeking to find the spatial distribution of high and low expression of distinct genes in the given 2D area.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
The Gestalt principle of similarity states that items alike in their properties (size, shape, color, and so on) tend to be perceived as being a related group. Here, by assigning spots that have similar levels of genes expressed with similar colors, I make it salient to see the patterns in expression levels. This grouping based on similar color also allows visualization of regions of high and low expression.
5. Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
library(ggplot2)
file <- "/Users/kevinnguyen/Desktop/genomic-data-visualization-2025/data/eevee.csv.gz"
data <- read.csv(file)
aligned_x <- data[, 2]
aligned_y <- data[, 3]
gexp <- data[, 5:ncol(data)]
gexp_tot <- rowSums(gexp != 0)
ggplot(data, aes(x = aligned_x, y = aligned_y)) +
geom_point(aes(color = gexp_tot),
size = 2,
alpha = 0.7) +
ggtitle("Spatial Distribution of Total Genes Expressed")+
labs(x = "X Position", y = "Y Position")+
scale_color_gradientn(
colors = c("blue","cyan","green","yellow","orange","red"),
name = "Total Genes Expressed"
) +
coord_fixed()
