Spatial Distribution of Cells by Nucleus-to-Cell Ratio Write Up
1. What data types are you visualizing?
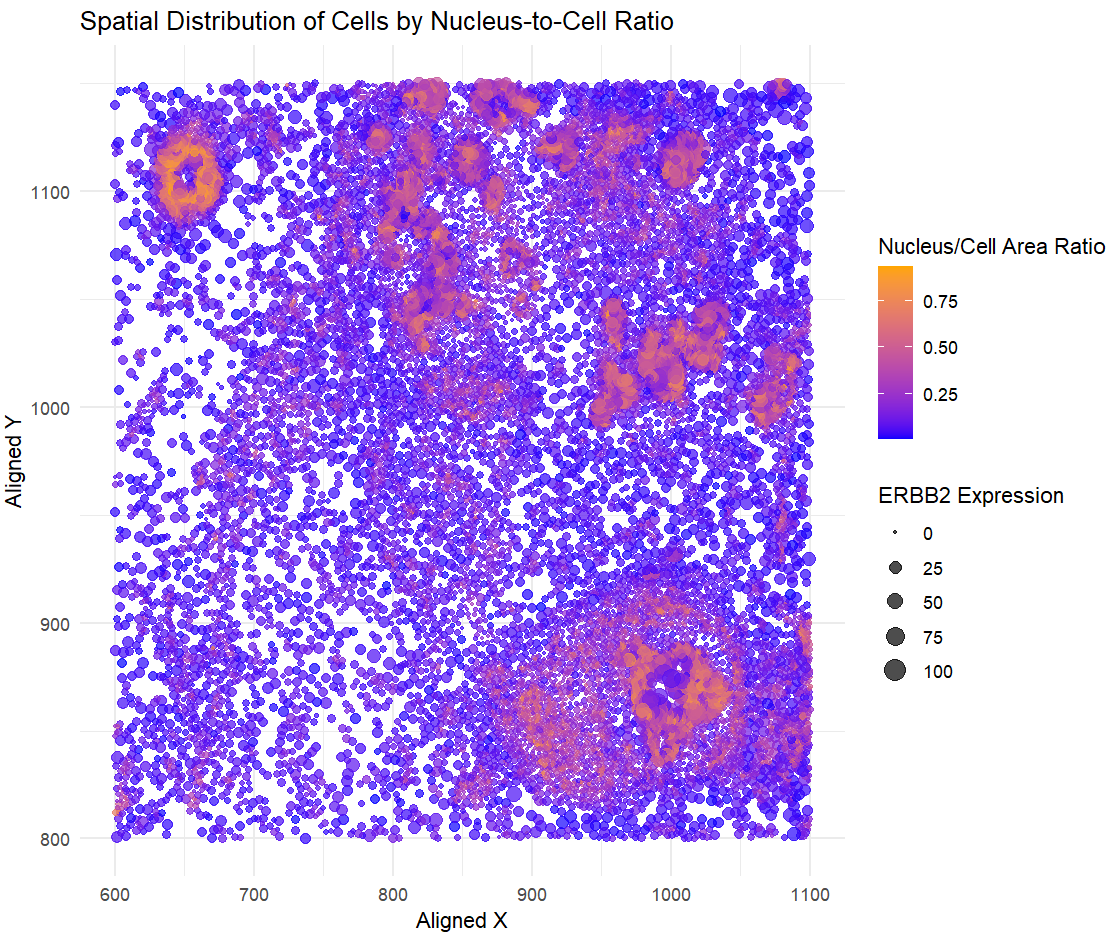
I am visualizing quantitative data representing the nucleus-to-cell area ratio for each cell, quantitative data of ERBB2 expression levels to indicate gene activity, and spatial data capturing the x, y centroid positions of each cell to maintain the tissue’s structural context.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I am using the geometric primitive of points to represent each cell. To encode the nucleus-to-cell area ratio, I am using the visual channel of color, ranging from blue (low ratio) to orange (high ratio). To encode the expression count of the ERBB2 gene, I am using the visual channel of size, where larger points indicate higher expression. To encode the spatial x position, I am using the visual channel of position along the x-axis, and to encode the spatial y position, I am using the visual channel of position along the y-axis to maintain the tissue’s structure.
3. What about the data are you trying to make salient through this data visualization?
My data visualization seeks to make more salient the relationship between the nucleus-to-cell area ratio and ERBB2 expression, while also preserving the spatial distribution of cells within the tissue. By encoding the nucleus-to-cell area ratio with color and ERBB2 expression with size, the visualization highlights potential correlations between cell morphology and gene expression patterns across different regions.
4. What Gestalt principles and/or knowledge about perceptiveness of visual encodings are you using to accomplish this?
I am using the Gestalt principle of similarity to enhance the perception of visual encodings in this plot. By using color to represent the nucleus-to-cell area ratio, cells with similar ratios appear visually grouped, making patterns and trends more intuitive to identify. This helps the viewer quickly discern clusters of cells with comparable morphology and how they relate to other encoded variables, improving overall readability and interpretation of the data.
5. Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
library(ggplot2)
file <- 'C:/Users/reach/OneDrive/Documents/2024-25/SPRING/Genomic Data Visualization/genomic-data-visualization-2025/data/pikachu.csv.gz'
data <- read.csv(file)
data[1:5, 1:10]
# Compute nucleus-to-cell area ratio
data$nucleus_ratio <- data$nucleus_area / data$cell_area
# Plot with nucleus ratio as color and ERBB2 expression as point size
ggplot(data) +
geom_point(aes(x = aligned_x,
y = aligned_y,
col = nucleus_ratio,
size = ERBB2),
alpha = 0.7) +
scale_color_gradient(low = "blue", high = "orange") +
scale_size_continuous(range = c(0.5, 5)) +
theme_minimal() +
labs(title = "Spatial Distribution of Cells by Nucleus-to-Cell Ratio",
x = "Aligned X",
y = "Aligned Y",
color = "Nucleus/Cell Area Ratio",
size = "ERBB2 Expression")
