Scatter Plot of POSTN vs LUM Expression
1. What data types are you visualizing?
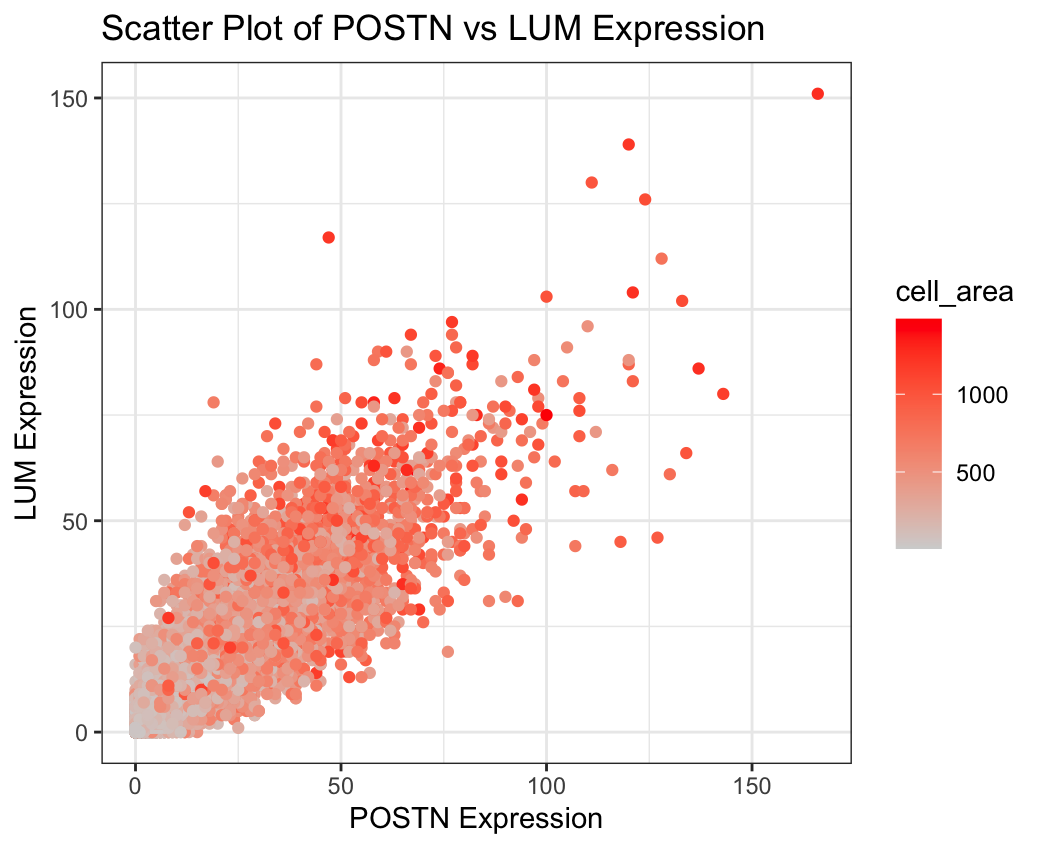
I am visualizing quantitative data for the expression levels of the POSTN and LUM genes in all the individual cells and I am also visualizing quantitative data for the cell areas which correspond to color encoded variables in my graph. POSTN expression is represented on the x axis and LUM expression is represented on the y axis and both are continuous numerical values. Cell area is color encoded and is also a continuous numerical value that represents the size of each cell in the data sample.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I am using the geometric primitive of points to encode each cell (shown as circles on the scatterplot). I used the visual channel of position along the x axis to encode the expression level of the POSTN gene and the visual channel of position along the y axis to encode expression of the LUM gene. I chose these two genes because they were the two most highly expressed genes. I also used the visual channel of color to encode the cell area, with a color change from grey to red as the physical cell size increased.
3. What about the data are you trying to make salient through this data visualization?
I was interested in examining if there was a relationship between POSTN and LUM gene expression across the individual cells because this could point to these genes being codependent. I chose these two specifically because when I summed up the gene expression columns for each gene and sorted the genes from highest to lowest gene expression across all cells, these genes were the two highest expressed. I also decided to add a color gradient to display the cell area to understand if the gene expression pattern and relationship between the two genes could depend on cell size.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
I used the idea of proximity because clusters suggest similarity and in my graph, the points clustered together suggest similar expression levels of POSTN and LUM. These clustered points could show some potential of co-expression. I also used the idea of similarity with my color encoding because this color gradient allows for pattern recognition and helps differentiate from small to large cell sizes. I also somewhat used continuity because the points follow a diagonal trend, so the correlation or trendline between POSTN and LUM expression can be seen and these points can be visualized as more of a continuous line.
5. Code (paste your code in between the ``` symbols)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
file <- '~/desktop/genomic-data-visualization-2025/data/pikachu.csv.gz'
data <- read.csv(file)
# remove first 6 cols and then find top 2 genes with greatest gene expression
gene_cols <- data[, 7:ncol(data)]
# sum expression for each gene across all cells and find the top two most expressed
gene_sums <- colSums(gene_cols)
top_genes <- names(sort(gene_sums, decreasing = TRUE))
top_genes
#top 2 are POSTN and LUM
ggplot(data) + geom_point(aes(x = POSTN,
y = LUM,
color = cell_area)) +
labs(title = "Scatter Plot of POSTN vs LUM Expression",
x = "POSTN Expression",
y = "LUM Expression") +
scale_color_gradient(low = 'lightgrey', high = 'red') +
theme_bw()
