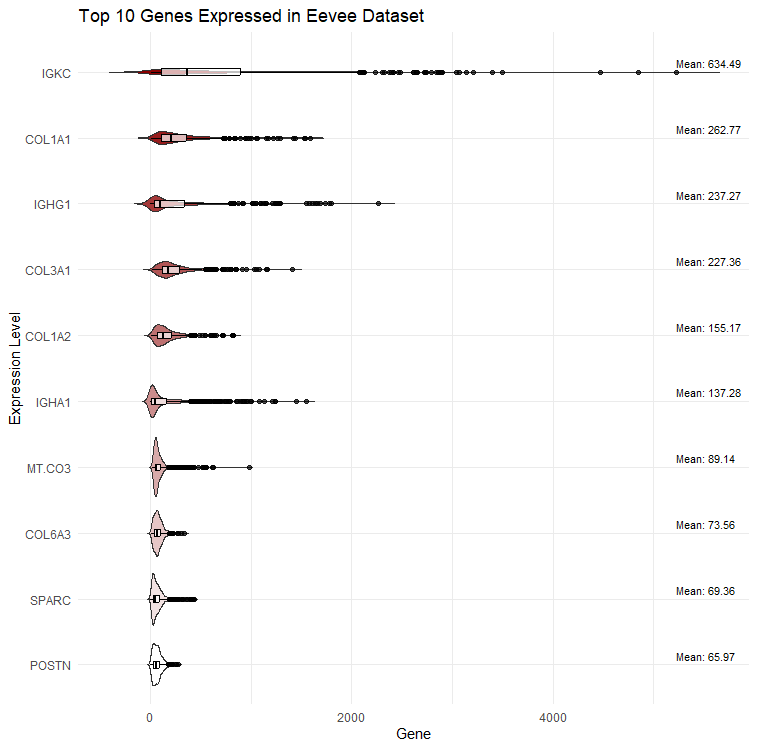
The Top 10 Genes Expressed in the Eevee Dataset
1. What data types are you visualizing?
I am visualizing gene expression data obtained through sequencing of the top 10 genes expressed in the Eevee dataset.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I am using 10 different colors as visual channels to describe the 10 genes of interest. The colors are arranged from dark red (highest gene expression) to white (lowest gene expression). To encode expression differences I am using the visual channel of size along the x-axis. Data labels are present to illustrate the large difference in expression between IGKC and the remaining gene cohort. Furthermore, by using both color and the text labels I am able to show subtle differences between violin plots that were not apparent previously.
The geometric primitives used in this dataset are points, where each point represents a cell. Areas are also used as geometric primitives where the (rough) area of the outer shape of the violin plot represents the distribution of a given cell’s gene expression data, which is estimated using the kernel density estimation. Wider sections (larger areas) indicate higher density.
3. What about the data are you trying to make salient through this data visualization?
My data visualization seeks to make more salient the expression levels and relative differences in the top 10 most highly expressed genes. While I tried other gene expression maps like volcano plots, I realized that the sheer number of genes present in the dataset detracted from the visualization, so I choose to focus on the top 10 through violin plots.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
I am using the gestalt principles of proximity and similarity because genes that are close in levels of expression to one another are next to each other and also similar in color. I am using color hues to illustrate the natural progression from most expressed genes to least expressed.
5. Code (paste your code in between the ``` symbols)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
library(ggplot2)
library(dplyr)
library(tidyr)
library(RColorBrewer)
file <- 'eevee.csv.gz'
data <- read.csv(file)
# determine the top 10 most highly expressed genes
top_10_genes <- names(sort(colMeans(data[, -(1:4)]), decreasing = TRUE)[1:10])
plot_data <- data %>%
select(all_of(top_10_genes)) %>%
pivot_longer(cols = everything(), names_to = "Gene", values_to = "Expression")
# mean expression calculation for each gene
mean_expression <- plot_data %>%
group_by(Gene) %>%
summarize(Mean = round(mean(Expression), 2)) %>%
arrange(desc(Mean))
plot_data$Gene <- factor(plot_data$Gene, levels = rev(mean_expression$Gene))
# create colors from dark red to white
color_palette <- colorRampPalette(c("darkred", "white"))(10)
# violin plot
violin_plot <- ggplot(plot_data, aes(x = Gene, y = Expression, fill = Gene)) +
geom_violin(trim = FALSE) +
geom_boxplot(width = 0.1, fill = "white", color = "black", alpha = 0.7) +
geom_text(data = mean_expression, aes(x = Gene, y = max(plot_data$Expression),
label = paste("Mean:", Mean)),
vjust = -0.5, hjust = 0, size = 3) +
theme_minimal() +
theme(axis.text.y = element_text(hjust = 1),
legend.position = "none") +
labs(title = "Top 10 Genes Expressed in Eevee Dataset",
x = "Expression Level",
y = "Gene") +
scale_fill_manual(values = rev(color_palette)) +
print(violin_plot)
