HW1: Gene expression pattern for GNB1 and HES4
1. What data types are you visualizing?
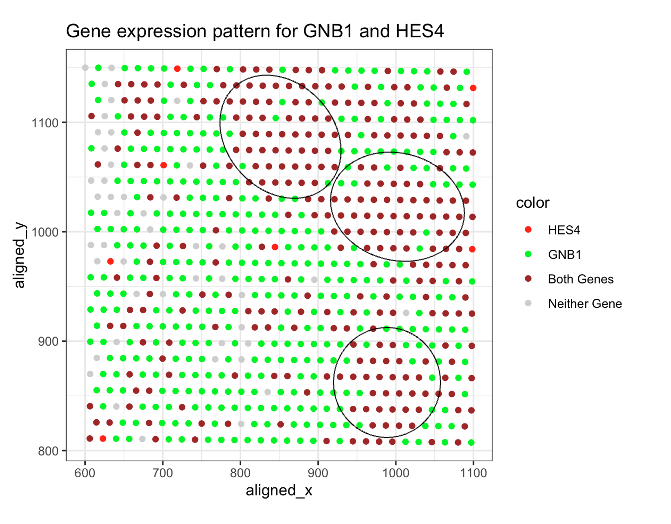
I am visualizing HES4 and GNB1’s spatial gene expression patterns for eevee.
2. What data encodings (geometric primitives and visual channels) are you using to visualize these data types?
I used points for genmetriic primitives.
I used color/hue, y-position and x-position for visual channels. I used these visual channcels because my data is categorical. I have different type of gene expression. And I want to show the position for these categorical data.
3. What about the data are you trying to make salient through this data visualization?
I am trying to use four different colors to demonstrate the spatial gene expression patterns for both genes. It is similar to creating a Venn diagram with the gene expressions. Red represents spots that only express HES4, green represents spots that only express GNB1, brown indicates spots where both genes are expressed, and light gray represents spots where neither gene is expressed.
I also want to highlight a specific pattern I observed: there are three regions with highly concentrated overlapping expressions of both HES4 and GNB1.
4. What Gestalt principles or knowledge about perceptiveness of visual encodings are you using to accomplish this?
I used similarity in color to group spots with same gene expression patterns, as shown in the legend.
Additionally, I used enclosures to highlight areas with high overlapping gene expressions
5. Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
library(ggplot2)
file <- "~/Downloads/eevee.csv.gz"
data <- read.csv(file)
data[1:5,1:10]
dim(data)
ncol(data)
nrow(data)
rownames(data)
colnames(data)
# visualize only HES4
ggplot(data) +
geom_point(aes(x = aligned_x, y = aligned_y, col = HES4)) +
scale_color_gradient(low = 'lightgrey', high = 'red')+
theme_bw()
# visualize only GNB1
ggplot(data) + geom_point(aes(x = aligned_x, y = aligned_y,col = GNB1)) +
scale_color_gradient(low = 'lightgrey',high = 'green')+
theme_bw()
# add a new color assignment column for each data point
data$color <- ifelse(
data$HES4 != 0 & data$GNB1 == 0, "red", ifelse(
data$HES4 == 0 & data$GNB1 != 0, "green", ifelse(
data$HES4 != 0 & data$GNB1 != 0, "brown",
"lightgrey"
)
)
)
# create a graph base on the new color assignment
ggplot(data) + geom_point(aes(x = aligned_x, y = aligned_y, col = color)) +
scale_color_identity(
guide = "legend",
labels = c("HES4", "GNB1", "Both Genes", "Neither Gene"),
breaks = c("red","green","brown","lightgrey")
) +
labs(title = "Gene expression pattern for GNB1 and HES4") + theme_bw()
