Visualizing the Impact of the Number of PCs used to perform Nonlinear Dimensionality Reduction using tSNE
Write a brief description of your figure so we know what you are visualizing.
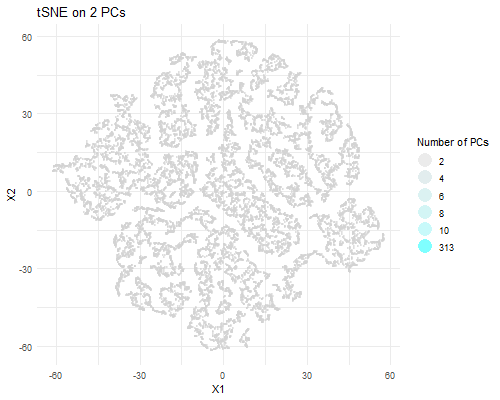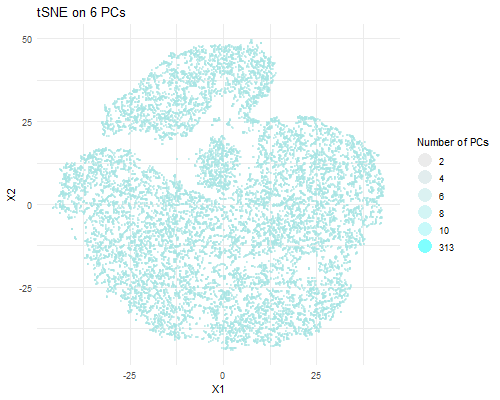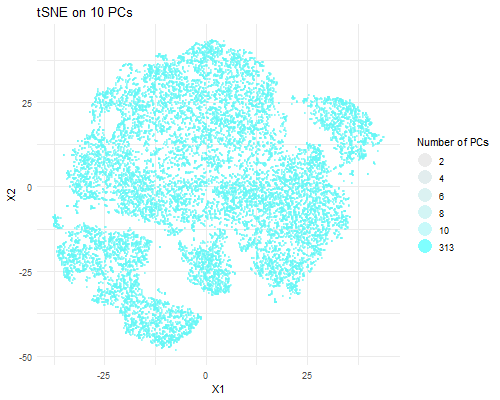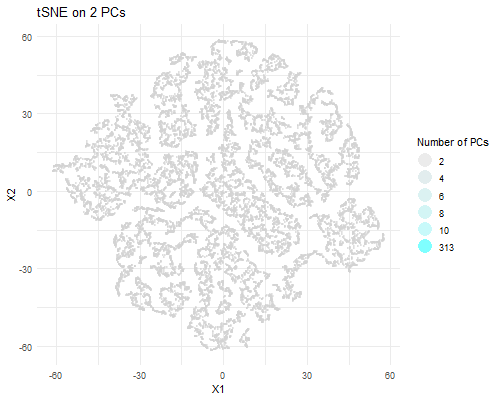
I am visualizing PCA data in tSNE space, animating the plots transitioning as the number of PCs I used in my calculation of the tSNE data rises (2, 4, 6, 8, 10, and 313).
If I perform non-linear dimensionality reduction on PCs, what happens when I vary how many PCs should I use?
As the number of PCs used increases, you can visualize more distinct clusters in tSNE space. When we plot tSNE with two PCs, we can’t really make out a distinct clusters. With four PCs, we can make out two clusters. With six PCs, we can make out three. With eight PCs, we can make out ~six distinct clusters. With 10, we can make out ~seven clusters. With all 313 PCs, we can make out a lot more clusters. This is likely due to the increase in the variance captured with a greater number of PCs. A greater number of PCs captures more information about the data, giving tSNE more info to work with and perform dimensionality reduction on and allowing it to form more local clusters in the data. Of course, that means that the tSNE data with the most useful information is the one which used all 313 PCs to perform dimensionality reduction.
Code
### Ayan Vaishnav
### Genomic Data Visualization 2025
### Homework Assignment Extra Credit 1
## Source for most of the code: Ayan's past HWs and Prof. Fan's code on gganimate
## Load libraries
library(ggplot2)
library(patchwork)
library(tidyverse)
library(Rtsne)
library(gganimate)
set.seed(6)
## Load Pikachu (imaging) dataset
file <- "~/Desktop/genomic-data-visualization-2025/data/pikachu.csv.gz"
data <- read.csv(file)
head(data)
names(data)
## Make smaller matrices
pos <- data[, 5:6]
rownames(pos) <- data$barcode
gexp <- data[, 7:ncol(data)]
rownames(gexp) <- data$barcode
gexp[1:10,1:10] # rows are cells, columns are genes
dim(gexp) # 17136 rows, 313 columns
## Normalize + log transform data
gexp_norm <- gexp / rowSums(gexp) # normalize!
gexp_norm <- gexp_norm * 10000
gexp_norm <- log10(gexp_norm + 1) # transform!
## Dimensionality reduction - first PCA, then tSNE on diff numbers of PCs
pca <- prcomp(gexp_norm)
emb2 <- Rtsne(pca$x[,1:2])
emb4 <- Rtsne(pca$x[,1:4])
emb6 <- Rtsne(pca$x[,1:6])
emb8 <- Rtsne(pca$x[,1:8])
emb10 <- Rtsne(pca$x[,1:10])
## Make data frames of the tSNE data
tsne_df_2_pcs <- data.frame(emb2$Y)
dim(tsne_df_2_pcs)
tsne_df_4_pcs <- data.frame(emb4$Y)
dim(tsne_df_4_pcs)
tsne_df_6_pcs <- data.frame(emb6$Y)
dim(tsne_df_6_pcs)
tsne_df_8_pcs <- data.frame(emb8$Y)
dim(tsne_df_8_pcs)
tsne_df_10_pcs <- data.frame(emb10$Y)
dim(tsne_df_10_pcs)
## Animation data frame
animate_df <- rbind(
cbind(tsne_df_2_pcs, order = 1),
cbind(tsne_df_4_pcs, order = 2),
cbind(tsne_df_6_pcs, order = 3),
cbind(tsne_df_8_pcs, order = 4),
cbind(tsne_df_10_pcs, order = 5)
)
## Plot
titles <- c("1" = "tSNE on 2 PCs", "2" = "tSNE on 4 PCs", "3" = "tSNE on 6 PCs", "4" = "tSNE on 8 PCs", "5" = "tSNE on 10 PCs")
p <- ggplot(animate_df, aes(x = X1, y = X2, col = order)) +
geom_point(size = 0.01, alpha = 0.5)
anim <- p +
transition_states(order) +
view_follow() +
ease_aes('linear') +
scale_color_gradient(low = "red", high = "blue", breaks = 1:5, labels = c("2", "4", "6", "8", "10")) +
labs(title = '{titles[as.character(closest_state)]}', color = "Number of PCs") +
guides(color = guide_legend(override.aes = list(size = 5, shape = 16, color = c("red", "#d5345d", "#ba2988", "#8f1dbd", "#0808f5")),
title = "Number of PCs")) +
theme_minimal()
## Animate
animate(anim, height = 400, width = 500)
