We are a bioinformatics research lab in the Center for Computational Biology and the Department of Biomedical Engineering at Johns Hopkins University. Meet our team!
We develop bioinformatics methods for analyzing spatially resolved sequencing and imaging data.
Spatial organization at both the subcellular-level within cells as well as the cellular-level within tissues play important roles in regulating cell identity and function. Recent technological advances have enabled high-throughput spatially resolved transcriptomic profiling at single-molecule and near-single-cell resolution. We develop machine learning and other statistical approaches as open-source computational software to take advantage of this new spatial information in deriving biological insights regarding how spatial organization plays a role in both healthy and diseased settings.
- Gohta Aihara, Kalen Clifton, Mayling Chen, Zhuoyan Li, Lyla Atta, Brendan F Miller, Rahul Satija, John W Hickey, Jean Fan^. SEraster - a rasterization preprocessing framework for scalable spatial omics data analysis. Bioinformatics. June 20, 2024. doi.org/10.1093/bioinformatics/btae412
- Lyla Atta, Kalen Clifton, Manjari Anant, Gohta Aihara, and Jean Fan^. Gene count normalization in single-cell imaging-based spatially resolved transcriptomics. Genome Biology. June 12, 2024. doi.org/10.1186/s13059-024-03303-w
- Kalen Clifton*, Manjari Anant*, Gohta Aihara, Lyla Atta, Osagie K Aimiuwu, Justus M Kebschull, Michael I Miller, Daniel Tward^, Jean Fan^. STalign - alignment of spatial transcriptomics data using diffeomorphic metric mapping. Nature Communications. December 8, 2023. doi.org/10.1038/s41467-023-43915-7
- Brendan F Miller, Feiyang Huang, Lyla Atta, Arpan Sahoo, Jean Fan^. Reference-free cell type deconvolution of pixel-resolution spatially resolved transcriptomics data. Nature Communications. 2022. doi:/10.1038/s41467-022-30033-z
- Lyla Atta, Jean Fan^. Computational challenges and opportunities in spatially resolved transcriptomic data analysis. Nature Communications. 2021. doi:10.1038/s41467-021-25557-9
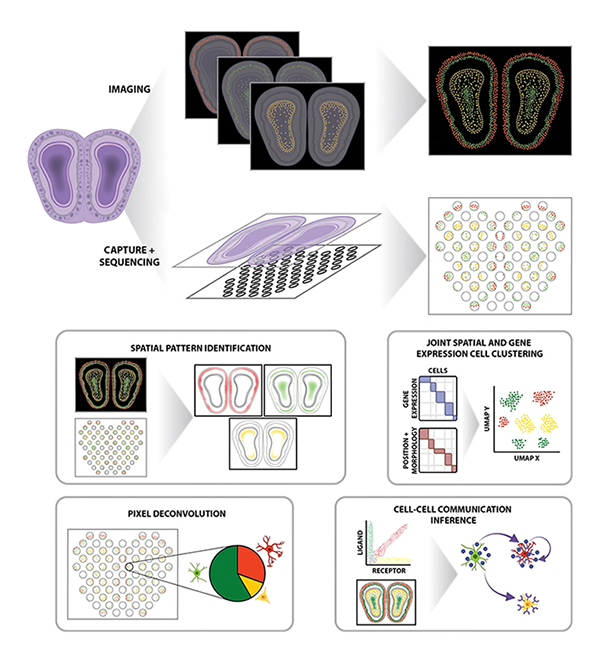
We apply these bioinformatics methods to better understand the role of cellular heterogeneity in disease.
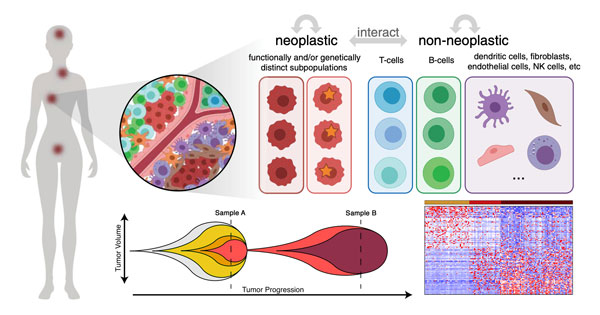
Advancements in high-throughput sequencing and imaging technologies have uncovered tremendous genetic, epigenetic, transcriptional, and spatial heterogeneity in various diseases but their impact on clinical outcomes is not well understood. We establish close collaborations with clinical collaborators to develop and apply bioinformatics methods that contribute to a more complete understanding of how cellular heterogeneity impacts disease progression and clinical prognosis.
- Jean Fan^, Kamil Slowikowski, Fan Zhang. Single-cell transcriptomics in cancer - computational challenges and opportunities. Nature Experimental and Molecular Medicine. 2020, doi.org:10.1038/s12276-020-0422-0
- Jean Fan*, Hae-Ock Lee*, Soohyun Lee, Da-eun Ryu, Semin Lee, et al. Linking transcriptional and genetic tumor heterogeneity through allele analysis of single-cell RNA-seq. Genome Research. 2018. doi:10.1101/gr.228080.117
- Lili Wang*, Jean Fan*, Joshua M. Francis, George Georghiou, Sarah Hergert, et al. Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Research. 2017. doi:10.1101/gr.217331.116