Querying Google Scholar with Rvest
Introduction
A number of posts on my social media feed has noted the rise of research papers containing obviously LLM-generated text, often using tools such as ChatGPT:

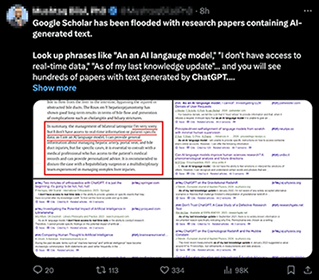


I sought to quantify the extent of the issue. So in this blog post, I will demonstrate how to use rvest, a web-scraping tool in R, to query Google Scholar to quantify the number of publications that contain obviously LLM-generated text. The code, of course, can be modified for other queries as well. Feel free to try it out for yourself!
Getting to know the structure of Google Scholar queries
Google Scholar is a web search engine that indexes research papers across an array of publishing formats and disciplines. Before we jump into using rvest, we need to first understand how we can automate querying Google Scholar.
As noted in the social media posts, LLM-generated text can contain signature phrases such as “as an AI language model”, “I don’t have access to real-time data”, “as of my last knowledge update” and so forth. I can use Google Scholar to query one of these phrases and quantify how many research papers that are index by Google Scholar contain this exact phrase.
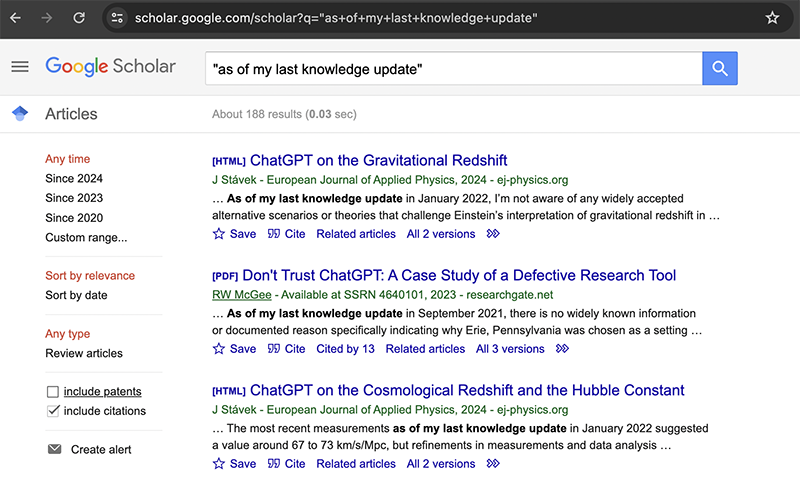
Many of these are legitimate research papers that are actually about ChatGPT though. I am specifically interested in counting the number of papers that contain LLM-generated text while not admitting to doing so. So I will exclude the word ‘ChatGPT’ from my query under the assumption that research papers attempting to hide their use of LLMs will not mention the model by name.
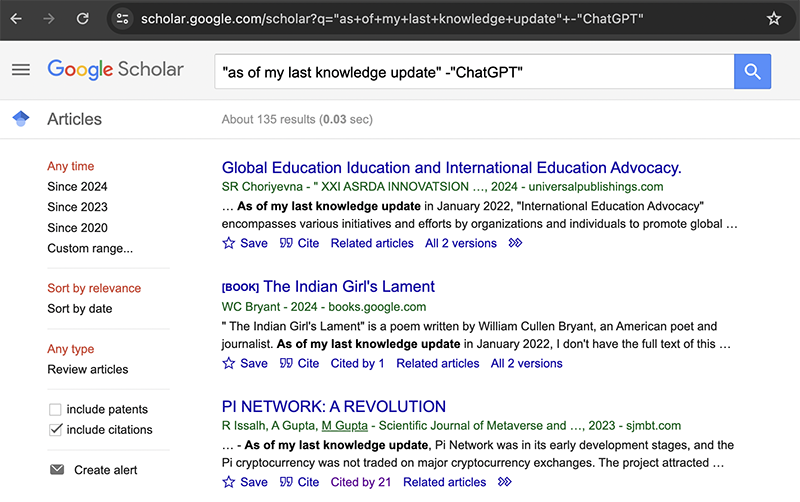
If I want to restrict to the research papers published between 2022 to 2023 for example, I can add a custom range.

So it looks like there are about 84 research papers that contain the exact phrase “as of my last knowledge update” but not “ChatGPT” published in the range of 2022 to 2023.
Note the structure of the URL:
https://scholar.google.com/scholar?q=%22as+of+my+last+knowledge+update%22+-%22ChatGPT%22&as_ylo=2022&as_yhi=2023
It contains the Google Scholar url with a query q, quotes %22, the query terms, the starting year as_ylo= and ending year as_yhi= This means, if we can use the URL to structure our query for different terms across different timespan. We will use this understanding to help us quantify the number of publications that contain a specific query text over time.
Web scraping with rvest
Given a URL, I can use rvest in R to scrape the HTML content.
library(rvest)
url <- "https://scholar.google.com/scholar?q=%22as+of+my+last+knowledge+update%22+-%22ChatGPT%22&as_ylo=2022&as_yhi=2023"
page <- read_html(url)
page
{html_document}
<html>
[1] <head>\n<title>Google Scholar</title>\n<meta http-equiv="Content-Type" content="text/html; charset=UTF-8">\n<meta http-equiv ...
[2] <body><div id="gs_top" onclick="">\n<style>#gs_md_s,.gs_md_wnw{z-index:1200;position:fixed;top:0;left:0;width:100%;height:10 ...
If I look at Google Scholar’s page source, I can see that the count of the number of results is in a div called gs_ab_mdw.
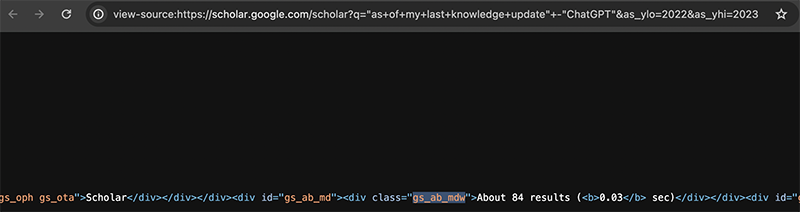
So I can use rvest to also grab that div node.
results <- page %>%
html_nodes(".gs_ab_mdw") %>%
html_text()
results
[1] "ArticlesScholar" "About 84 results (0.02 sec)"
It looks like what I want corresponds to the second element of the list that is returned. With a little regex, I can grab the number associated. I will also delete any commas and cast to a numeric for later.
numpubs <- stringr::str_extract(results[[2]], "\\d{1,3}(,\\d{3})*")
as.numeric(gsub(",", "", numpubs))
[1] 84
Putting it all together
Putting everything together, I will loop through a set of years ranging from 2018 to 2024 and query Google Scholar for the exact phrase “as+of+my+last+knowledge+update”, scrape the HTML, and extract the estimated count of publications fitting those query specifications.
years <- seq(2018, 2024)
querytext <- "as+of+my+last+knowledge+update"
numpubs <- sapply(years, function(year) {
url <- paste0("https://scholar.google.com/scholar?q=%22", querytext, "%22+-%22ChatGPT%22&as_ylo=", year, "&as_yhi=", year+1)
page <- read_html(url)
results <- page %>%
html_nodes(".gs_ab_mdw") %>%
html_text()
numpubs <- stringr::str_extract(results[[2]], "\\d{1,3}(,\\d{3})*")
as.numeric(gsub(",", "", numpubs))
})
numpubs
[1] 4 6 7 9 84 115 35
What if I use a different query text?
querytext <- "as+an+AI+language+model"
numpubs2 <- sapply(years, function(year) {
url <- paste0("https://scholar.google.com/scholar?q=%22", querytext, "%22+-%22ChatGPT%22&as_ylo=", year, "&as_yhi=", year+1)
page <- read_html(url)
results <- page %>%
html_nodes(".gs_ab_mdw") %>%
html_text()
numpubs <- stringr::str_extract(results[[2]], "\\d{1,3}(,\\d{3})*")
as.numeric(gsub(",", "", numpubs))
})
numpubs2
[1] 1 1 5 10 61 63 8
How does this compare to the number of research articles that contain legitimate phrases such as “spatial transcriptomics”?
querytext <- "spatial+transcriptomics"
numpubs3 <- sapply(years, function(year) {
url <- paste0("https://scholar.google.com/scholar?q=%22", querytext, "%22+-%22ChatGPT%22&as_ylo=", year, "&as_yhi=", year+1)
page <- read_html(url)
results <- page %>%
html_nodes(".gs_ab_mdw") %>%
html_text()
numpubs <- stringr::str_extract(results[[2]], "\\d{1,3}(,\\d{3})*")
as.numeric(gsub(",", "", numpubs))
})
numpubs3
[1] 743 1370 3190 5840 9540 7550 1600
Or “single cell transcriptomics”?
querytext <- "single+cell+transcriptomics"
numpubs4 <- sapply(years, function(year) {
url <- paste0("https://scholar.google.com/scholar?q=%22", querytext, "%22+-%22ChatGPT%22&as_ylo=", year, "&as_yhi=", year+1)
page <- read_html(url)
results <- page %>%
html_nodes(".gs_ab_mdw") %>%
html_text()
numpubs <- stringr::str_extract(results[[2]], "\\d{1,3}(,\\d{3})*")
as.numeric(gsub(",", "", numpubs))
})
numpubs4
[1] 5310 8430 12900 15700 19900 13500 2800
Or “transcriptomics” in general?
querytext <- "transcriptomics"
numpubs5 <- sapply(years, function(year) {
url <- paste0("https://scholar.google.com/scholar?q=%22", querytext, "%22+-%22ChatGPT%22&as_ylo=", year, "&as_yhi=", year+1)
page <- read_html(url)
results <- page %>%
html_nodes(".gs_ab_mdw") %>%
html_text()
numpubs <- stringr::str_extract(results[[2]], "\\d{1,3}(,\\d{3})*")
as.numeric(gsub(",", "", numpubs))
})
numpubs5
[1] 65600 32000 73600 27500 51900 21100 14000
Given that we are in R, let’s use ggplot2 and gganimate to visualize all these trends together!
library(ggplot2)
library(gganimate)
df <- data.frame(rbind(
cbind(years, numpubs=numpubs, query="as+of+my+last+knowledge+update"),
cbind(years, numpubs=numpubs2, query="as+an+AI+language+model"),
cbind(years, numpubs=numpubs3, query="spatial+transcriptomics"),
cbind(years, numpubs=numpubs4, query="single+cell+transcriptomics"),
cbind(years, numpubs=numpubs5, query="transcriptomics")
))
df$years <- as.numeric(df$years)
df$numpubs <- as.numeric(df$numpubs)
df$ind <- 1:nrow(df)
p <- ggplot(df, aes(x = years, y = numpubs, col=query)) +
geom_line() +
theme_bw()
anim <- p +
view_follow(fixed_x = TRUE) +
geom_point() +
transition_reveal(ind)
anim
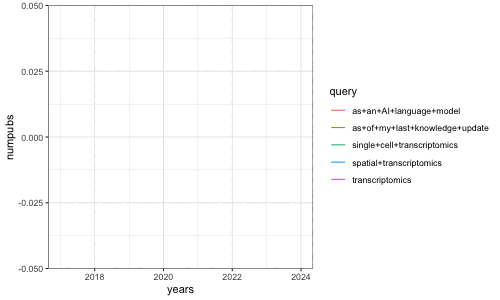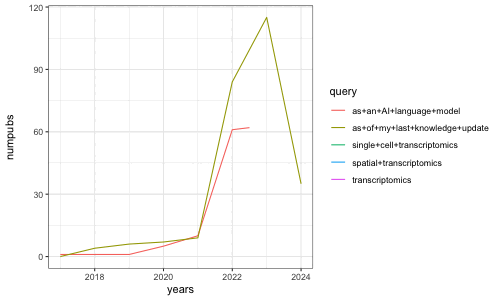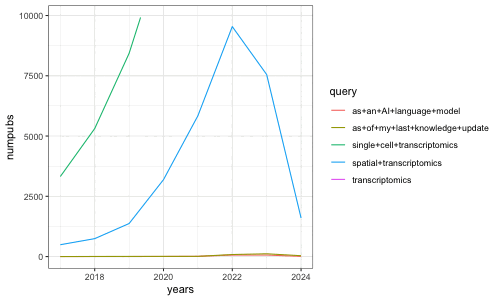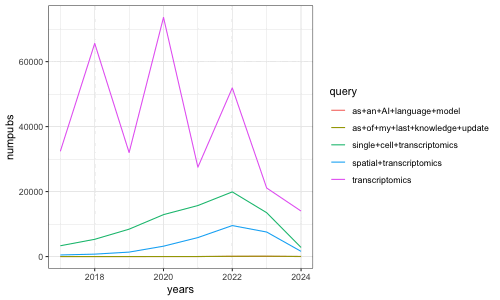
Indeed, we see a clear ‘boom’ or rise in the number of research papers containing obviously LLM-generated text in 2022 when these models were made more broadly accessible. Of course, we’re only a few months into 2024 so the number of publications in 2024 currently represents only these few months while all other years represent the whole year. Likewise, of course we are only identifying research papers containing obvious LLM-generated text based on perfect string matching, which presents limitations. More sophisticated approaches for identifying text likely to be substantially modified or produced by LLMs can be found in works such as Liang et al, “Monitoring AI-Modified Content at Scale: A Case Study on the Impact of ChatGPT on AI Conference Peer Reviews”, arXiv 2024 for example.
Final thoughts
While it is indeed true that at least 100s of published research papers that are just copy-pasted content from LLM models, they represent a very small percent of the total research that is published.
In my opinion, the existence of these papers brings to light a deeper, systemic issue regarding the publish-or-perish pressure that academics may feel, often under time pressure with limited resources, leading some to such dubious use of LLM models. As such, the prevelance of research papers containing obviously LLM-generated text may warrant further monitoring as well a deeper reflection by institutions, funding agencies, peer reviewers, and researchers themselves on potential mechanisms for promoting a culture of integrity in scientific publishing and enforcing consequences when such integrity is violated.
PS: If you do try to run this code too many times, you may encounter a temporary error “Response code 429. Google is rate limiting you for making too many requests too quickly.”
Recent Posts
- The Mentorship Index on 01 March 2026
- Vibe coding with SEraster and STcompare to compare spatial transcriptomics technologies on 22 February 2026
- RNA velocity in situ infers gene expression dynamics using spatial transcriptomics data on 13 October 2025
- Analyzing ICE Arrest Data - Part 2 on 27 September 2025
- Analyzing ICE Detention Data from 2021 to 2025 on 10 July 2025