Story-telling with Data Visualization
Aug 12, 2021
Introduction
In our last blog post, we used our recently published bioinformatics tool MERINGUE to identify spatial gene expression patterns in the new MERFISH spatially resolved transcriptomics data of the mouse cortex from Vizgen.
Data visualization remains an important hypothesis generating and analytical technique in data-driven research to facilitate new discoveries. As such, visualizing these identified gene expression patterns is a natural and intuitive way to communicate certain results such as regarding the association between spatial gene expression patterns and the underlying spatial organization of cell-types.
However, must we be limited to static visualizations? Animations can help draw attention to certain aspects of the data, in particular how datapoints are related across different visualizations, and enhance our understanding of the data. Though scientific journals are limited to static pictures (for now), perhaps we can begin integrating more dynamic data visualizations into our presentations, or even blog posts to enhance communication with the audience.
If a static picture is worth a thousand words, perhaps an animation is worth a million?
Creating animated data visualizations with gganimate
We previously discussed how we can use the gganimate package to visualize cell cycle genes. Here, I will use gganimate again to visually communicate the following points:
- spatially resolved transcriptomics allow us to measure gene expression within cells while preserving spatial context within tissues
- these cells can be clustered into transcriptionally distinct cell-types based on their measured gene expression
- these cell-types may be spatially organized within tissues
- for a given cell-type, genes may exhibit spatially organized expression patterns
library(gganimate)
## start from where the last tutorial left off
## put on same scale for animation purposes
emb <- scale(emb)
pos <- scale(pos)
range(emb)
range(pos)
## pick and animate a few genes
library(Matrix)
genes.final <- c('Slc17a7', "Epha6", 'Gad1', "Peg10", 'Gjc3')
length(genes.final)
m <- mat[genes.final,]
m <- t(scale(t(m)))
range(m)
m[m > 1] <- 1
m[m < -1] <- -1
dim(m)
df.all <- do.call(rbind, lapply(1:length(genes.final), function(i) {
gexp <- m[genes.final[i], rownames(pos)]
df <- data.frame(pos, gexp, gene=genes.final[i], order=i)
}))
p <- ggplot(df.all, aes(x = center_x, y = center_y, col=gexp)) +
geom_point(size = 0.1, alpha=0.1)
p <- p + theme_void() +
theme(legend.position = "none") +
scale_color_distiller(palette = 'RdBu',
limits = c(-1,1)) +
xlim(-3, 3) + ylim(-3,3)
anim1 <- p +
transition_states(order,
transition_length = 5,
state_length = 5) +
labs(title = '{genes.final[as.integer(closest_state)]}') +
theme(plot.title = element_text(size = 28)) +
enter_fade()
anim1
## visualize clustering
a <- data.frame(emb, com, 'umap')
b <- data.frame(pos, com, 'pos')
colnames(a) <- colnames(b) <- c('x', 'y', 'com', 'type')
df.trans <- rbind(a, b)
p <- ggplot(df.trans, aes(x = x, y = y)) +
geom_point(size = 0.1, alpha=0.1, show.legend = FALSE) +
theme_void() +
xlim(-3, 3) + ylim(-3,3)
anim2 <- p +
transition_states(type,
transition_length = 5,
state_length = 1) +
labs(title = '{closest_state}') +
theme(plot.title = element_text(size = 28)) +
enter_fade()
anim2
## color by clusters
p <- ggplot(df.trans, aes(x = x, y = y, col=com)) +
geom_point(size = 0.1, alpha=0.1, show.legend = FALSE) +
theme_void() +
xlim(-3, 3) + ylim(-3,3)
anim3 <- p +
transition_states(type,
transition_length = 5,
state_length = 1) +
labs(title = '{closest_state}') +
theme(plot.title = element_text(size = 28)) +
enter_fade()
anim3
## focus on one cluster
ct = 12 ## pyrimidal layer
df.12 <- data.frame(pos, com==ct, 'pos')
colnames(df.12) <- c('x', 'y', 'com', 'type')
head(df.12)
p <- ggplot(df.12, aes(x = x, y = y, col=com)) +
geom_point(size = 0.1, alpha=0.1, show.legend = FALSE) +
scale_color_manual(values=c("#dddddd", "#E69F00")) +
theme_void() +
xlim(-3, 3) + ylim(-3,3)
p + labs(title = 'cluster 12') +
theme(plot.title = element_text(size = 28))
## animate cluster 12 genes
## previously found by MERINGUE
genes.final12 <- c('Gpr161', 'Epha10', 'Amigo2','Mas1','Kit', 'Ephb1')
m <- mat[genes.final12,cells]
m <- t(scale(t(m)))
range(m)
m[m > 1] <- 1
m[m < -1] <- -1
df.all <- do.call(rbind, lapply(1:length(genes.final), function(i) {
gexp <- m[genes.final12[i], ]
gexp <- gexp[rownames(pos)]
names(gexp) <- rownames(pos)
df <- data.frame(pos, gexp, gene=genes.final[i], order=i)
}))
p <- ggplot(df.all, aes(x = center_x, y = center_y, col=gexp)) +
geom_point(size = 0.1, alpha=0.1)
p <- p + theme_void() +
theme(legend.position = "none") +
scale_color_distiller(palette = 'RdBu',
limits = c(-1,1),
na.value="#dddddd") +
xlim(-3, 3) + ylim(-3,3)
anim4 <- p +
transition_states(order,
transition_length = 5,
state_length = 5) +
labs(title = 'MERINGUE: {genes.final12[as.integer(closest_state)]}') +
theme(plot.title = element_text(size = 28)) +
enter_fade()
anim4
Stitching all 4 animations together:
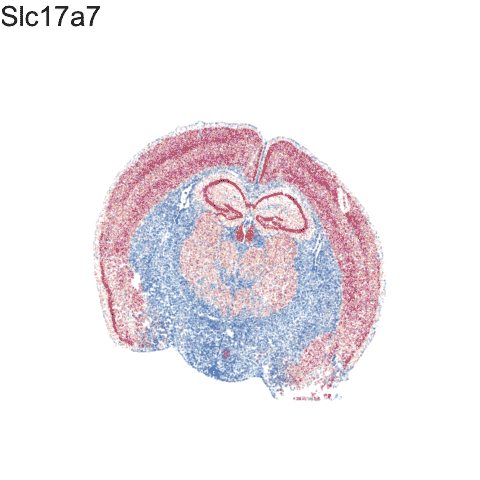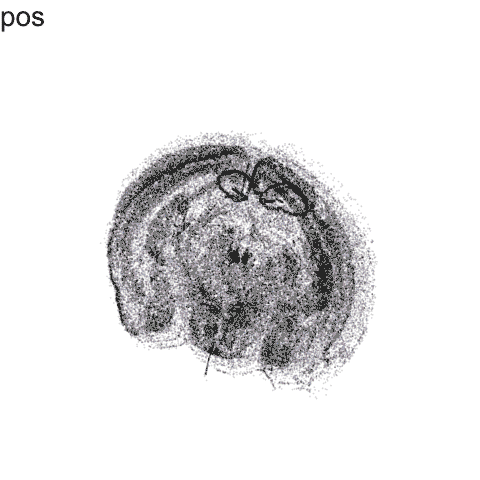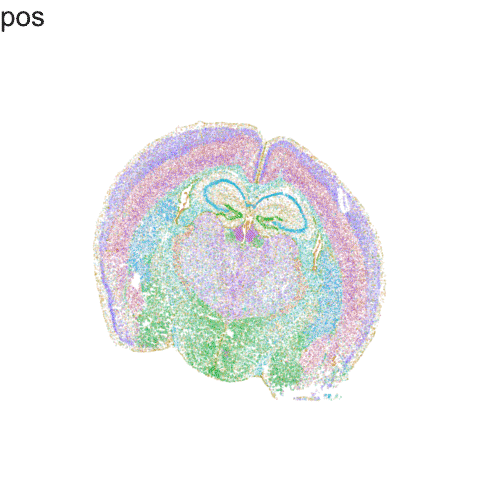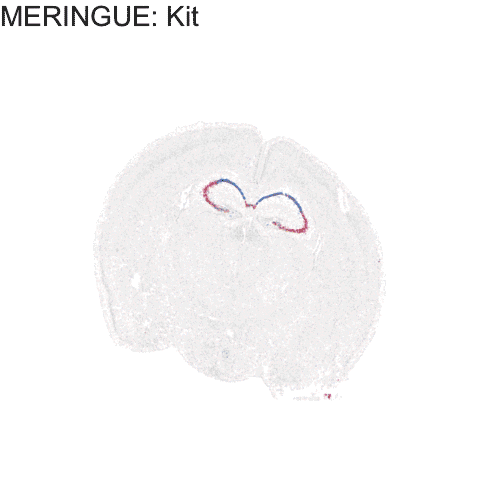
Try it out for yourself!
- Given this series of animations, what do you think is being communicated?
- Do you think this animation is effective at communicating the above points?
- What additional data visualizations can be integrated to more effectively communicate the above points?
- Check out gganimate.com for more
RECENT POSTS
- The many ways to calculate Moran's I for identifying spatially variable genes in spatial transcriptomics data on 29 August 2024
- Characterizing spatial heterogeneity using spatial bootstrapping with SEraster on 23 July 2024
- I use R to (try to) figure out which hospital I should go to for shoppable medical services by comparing costs through analyzing Hospital Price Transparency data on 22 April 2024
- Cross modality image alignment at single cell resolution with STalign on 11 April 2024
- Spatial Transcriptomics Analysis Of Xenium Lymph Node on 24 March 2024
- Querying Google Scholar with Rvest on 18 March 2024
- Alignment of Xenium and Visium spatial transcriptomics data using STalign on 27 December 2023
- Aligning 10X Visium spatial transcriptomics datasets using STalign with Reticulate in R on 05 November 2023
- Aligning single-cell spatial transcriptomics datasets simulated with non-linear disortions on 20 August 2023
- 10x Visium spatial transcriptomics data analysis with STdeconvolve in R on 29 May 2023
TAGS
RELATED POSTS
- The many ways to calculate Moran's I for identifying spatially variable genes in spatial transcriptomics data
- Characterizing spatial heterogeneity using spatial bootstrapping with SEraster
- I use R to (try to) figure out which hospital I should go to for shoppable medical services by comparing costs through analyzing Hospital Price Transparency data
- Cross modality image alignment at single cell resolution with STalign