Exploring UMAP parameters in visualizing single-cell spatially resolved transcriptomics data
Jan 19, 2022
Introduction
A student learning how to do single-cell and spatial transcriptomics analysis recently remarked how nice my UMAPs looked compared to theirs. I was surprised since my sense was that our filtering, normalization, and other processing of the data was quite similar. The main source of the difference turned out to be the different default parameters being used in the different UMAP implementations we were using. So in this blog post, I explore how different UMAP parameters can influence visualization of single-cell spatially resolved transcriptomics data.
Getting Started
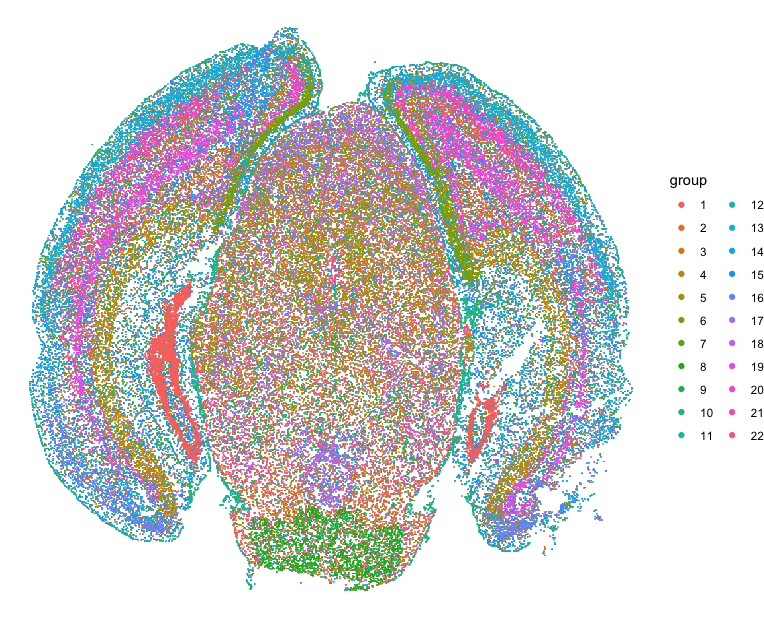
Let’s first read in the data and perform some single-cell transcriptional clustering on the MERFISH data of the mouse cortex from Vizgen.
## read in data
cd <- read.csv('~/Desktop/mouse_cortex_analysis/data/datasets_mouse_brain_map_BrainReceptorShowcase_Slice1_Replicate1_cell_by_gene_S1R1.csv.gz', row.names = 1)
annot <- read.csv('~/Desktop/mouse_cortex_analysis/data/datasets_mouse_brain_map_BrainReceptorShowcase_Slice1_Replicate1_cell_metadata_S1R1.csv.gz', row.names = 1)
pos <- annot[, c('center_x', 'center_y')]
pos[,2] <- -pos[,2] ## flip Y coordinates
library(MERINGUE)
## remove blanks
good.genes <- colnames(cd)[!grepl('Blank', colnames(cd))]
counts <- t(cd[, good.genes])
## filter
counts <- MERINGUE::cleanCounts(counts, verbose = FALSE)
## normalize, log transform
mat <- MERINGUE::normalizeCounts(counts, log = TRUE)
dim(mat)
## pca, take top 30 PCs
pca <- RSpectra::svds(A = t(mat), k=30,
opts = list(center = TRUE, scale = FALSE, maxitr = 2000, tol = 1e-10))
pcs <- as.matrix(t(mat) %*% pca$v[,1:30])
## subset data for clustering and impute rest to make faster
com <- MUDAN::getApproxComMembership(pcs, k=10,
nsubsample = nrow(pcs)*0.1,
method=igraph::cluster_louvain)
## plot clusters on original spatial positions
library(ggplot2)
library(scattermore)
d <- data.frame(x=pos$center_x, y=pos$center_y, group=com)
p <- ggplot(d) +
geom_scattermore(
mapping = aes(x = x, y = y, color = group),
pointsize = 1,
pixels = c(1000, 1000)
) +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.title = element_blank(),
legend.position = "none"
)
p <- p + theme_void()
p
Exploring UMAP parameters
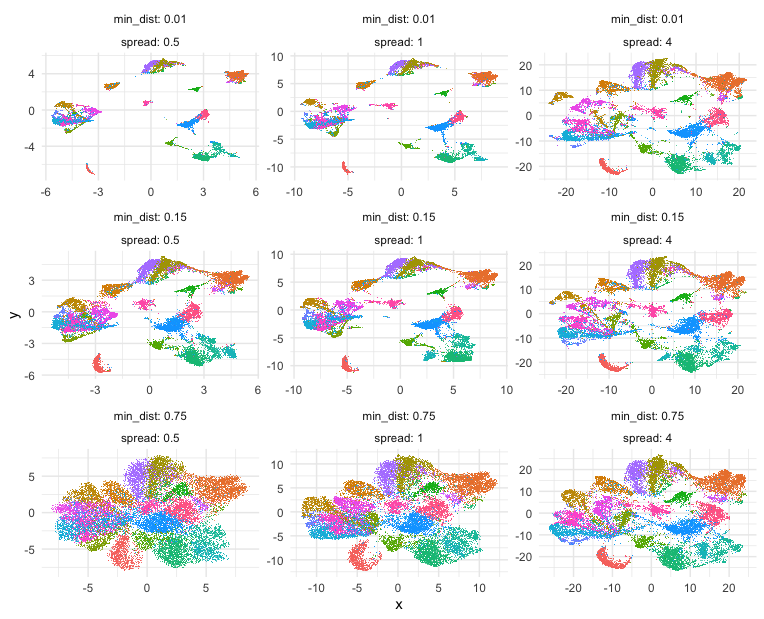
Building on some code courtesy of Kamil Slowikowski’s Gist, let’s try out some different UMAP parameters, namely min_dist and spread, and see how they impact the final 2D UMAP embedding and visualization.
## try some different parameters
umap_params <- expand.grid(
spread = c(0.5, 1, 4),
min_dist = c(0.01, 0.15, 0.75)
)
umaps <- lapply(seq(nrow(umap_params)), function(i) {
print(i)
emb <- uwot::umap(
X = pcs,
spread = umap_params$spread[i],
min_dist = umap_params$min_dist[i],
)
rownames(emb) <- colnames(mat)
return(emb)
})
library(data.table)
d <- rbindlist(lapply(seq(nrow(umap_params)), function(i) {
data.table(
x = umaps[[i]][,1],
y = umaps[[i]][,2],
spread = umap_params$spread[i],
min_dist = umap_params$min_dist[i],
group = com
)
}))
library(ggplot2)
library(scattermore)
p <- ggplot(d) +
geom_scattermore(
mapping = aes(x = x, y = y, color = group),
pointsize = 1,
pixels = c(1000, 1000)
) +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.title = element_blank(),
legend.position = "none"
) +
facet_wrap(min_dist ~ spread ,
labeller = label_both,
scales = "free")
p <- p + theme_minimal() +
theme(legend.position = "none")
Indeed, different combinations of parameters may give us somewhat different impressions of how different these transcriptionally distinct clusters of cells are!
Just for fun
Just for fun, let’s take what we learned from Story-telling with Data Visualization and animate two of the UMAP embeddings to see how the cells transition between these embeddings.
## animation
a <- data.frame(umaps[[1]], com, 'umap (min_dist: 0.01, spread: 0.5)')
b <- data.frame(umaps[[9]], com, 'umap (min_dist: 0.75, spread: 4)')
colnames(a) <- colnames(b) <- c('x', 'y', 'group', 'type')
df.trans <- rbind(a, b)
p <- ggplot(df.trans, aes(x = x, y = y)) +
geom_scattermore(
mapping = aes(x = x, y = y, color = group),
pointsize = 1,
pixels = c(1000, 1000)
)
anim <- p +
transition_states(type,
transition_length = 5,
state_length = 1) +
labs(title = '{closest_state}') +
theme(plot.title = element_text(size = 28)) +
enter_fade()
anim + theme_minimal() +
theme(legend.position = "none") +
view_follow()
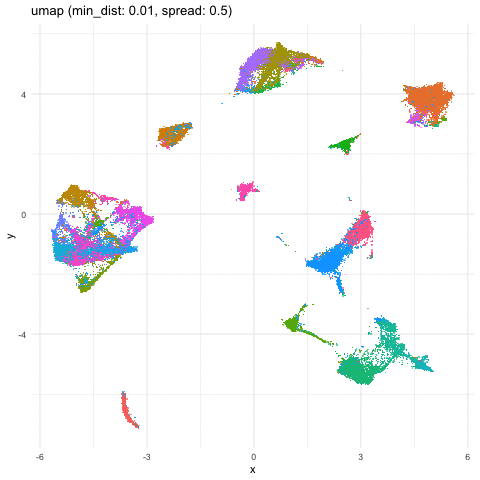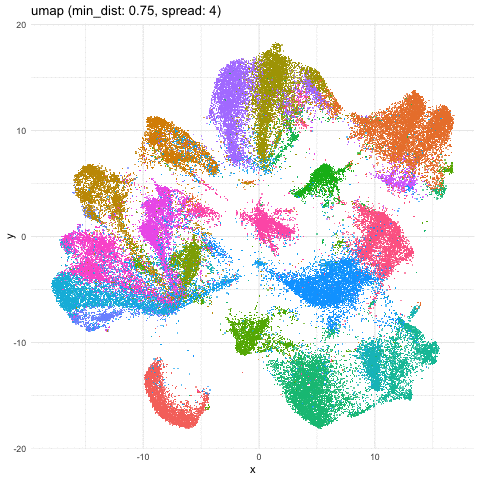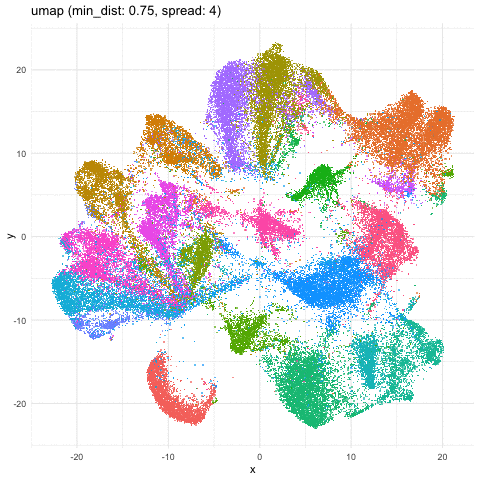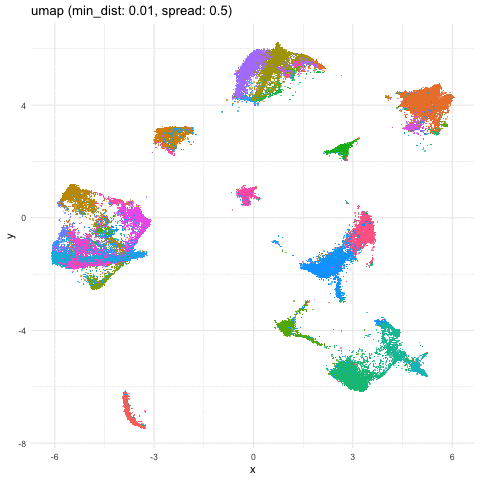
A good reminder to always follow up on the hunches a visualization gives you and dig into what are the genes driving these different cell clusters. Ultimately, we should do our best in interpreting these visualizations in the context of the underlying data!
Try it out for yourself!
- What are some other parameters we can toggle? Check out the original UMAP paper or documentation for ideas.
- Try out a different dimensionality reduction approach like tSNE
- What if we vary other things the number of PCs?
RECENT POSTS
- Characterizing spatial heterogeneity using spatial bootstrapping with SEraster on 23 July 2024
- I use R to (try to) figure out which hospital I should go to for shoppable medical services by comparing costs through analyzing Hospital Price Transparency data on 22 April 2024
- Cross modality image alignment at single cell resolution with STalign on 11 April 2024
- Spatial Transcriptomics Analysis Of Xenium Lymph Node on 24 March 2024
- Querying Google Scholar with Rvest on 18 March 2024
- Alignment of Xenium and Visium spatial transcriptomics data using STalign on 27 December 2023
- Aligning 10X Visium spatial transcriptomics datasets using STalign with Reticulate in R on 05 November 2023
- Aligning single-cell spatial transcriptomics datasets simulated with non-linear disortions on 20 August 2023
- 10x Visium spatial transcriptomics data analysis with STdeconvolve in R on 29 May 2023
- Impact of normalizing spatial transcriptomics data in dimensionality reduction and clustering versus deconvolution analysis with STdeconvolve on 04 May 2023
TAGS
RELATED POSTS
- Characterizing spatial heterogeneity using spatial bootstrapping with SEraster
- I use R to (try to) figure out which hospital I should go to for shoppable medical services by comparing costs through analyzing Hospital Price Transparency data
- Cross modality image alignment at single cell resolution with STalign
- Spatial Transcriptomics Analysis Of Xenium Lymph Node